Quantitative Fluorescence Microscopy of Protein Dynamics in Living Cells
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Two-Photon Excitation, Fluorescence Microscopy, and Quantitative Measurement of Two-Photon Absorption Cross Sections
Portland State University PDXScholar Dissertations and Theses Dissertations and Theses Fall 12-1-2017 Two-Photon Excitation, Fluorescence Microscopy, and Quantitative Measurement of Two-Photon Absorption Cross Sections Fredrick Michael DeArmond Portland State University Let us know how access to this document benefits ouy . Follow this and additional works at: https://pdxscholar.library.pdx.edu/open_access_etds Part of the Physics Commons Recommended Citation DeArmond, Fredrick Michael, "Two-Photon Excitation, Fluorescence Microscopy, and Quantitative Measurement of Two-Photon Absorption Cross Sections" (2017). Dissertations and Theses. Paper 4036. 10.15760/etd.5920 This Dissertation is brought to you for free and open access. It has been accepted for inclusion in Dissertations and Theses by an authorized administrator of PDXScholar. For more information, please contact [email protected]. Two-Photon Excitation, Fluorescence Microscopy, and Quantitative Measurement of Two-Photon Absorption Cross Sections by Fredrick Michael DeArmond A dissertation submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy in Applied Physics Dissertation Committee: Erik J. Sánchez, Chair Erik Bodegom Ralf Widenhorn Robert Strongin Portland State University 2017 ABSTRACT As optical microscopy techniques continue to improve, most notably the development of super-resolution optical microscopy which garnered the Nobel Prize in Chemistry in 2014, renewed emphasis has been placed on the development and use of fluorescence microscopy techniques. Of particular note is a renewed interest in multiphoton excitation due to a number of inherent properties of the technique including simplified optical filtering, increased sample penetration, and inherently confocal operation. With this renewed interest in multiphoton fluorescence microscopy, comes increased interest in and demand for robust non-linear fluorescent markers, and characterization of the associated tool set. -

Second Harmonic Imaging Microscopy
170 Microsc Microanal 9(Suppl 2), 2003 DOI: 10.1017/S143192760344066X Copyright 2003 Microscopy Society of America Second Harmonic Imaging Microscopy Leslie M. Loew,* Andrew C. Millard,* Paul J. Campagnola,* William A. Mohler,* and Aaron Lewis‡ * Center for Biomedical Imaging Technology, University of Connecticut Health Center, Farmington, CT 06030-1507 USA ‡ Division of Applied Physics, Hebrew University of Jerusalem, Jerusalem 91904, Israel Second Harmonic Generation (SHG) has been developed in our laboratories as a high- resolution non-linear optical imaging microscopy (“SHIM”) for cellular membranes and intact tissues. SHG is a non-linear process that produces a frequency doubling of the intense laser field impinging on a material with a high second order susceptibility. It shares many of the advantageous features for microscopy of another more established non-linear optical technique: two-photon excited fluorescence (TPEF). Both are capable of optical sectioning to produce 3D images of thick specimens and both result in less photodamage to living tissue than confocal microscopy. SHG is complementary to TPEF in that it uses a different contrast mechanism and is most easily detected in the transmitted light optical path. It also does not arise via photon emission from molecular excited states, as do both 1- and 2-photon excited fluorescence. SHG of intrinsic highly ordered biological structures such as collagen has been known for some time but only recently has the full potential of high resolution 3D SHIM been demonstrated on live cells and tissues. For example, Figure 1 shows SHIM from microtubules in a living organism, C. elegans. The images were obtained from a transgenic nematode that expresses a ß-tubulin-green fluorescent protein fusion and Figure 1 also shows the TPEF image from this molecule for comparison. -
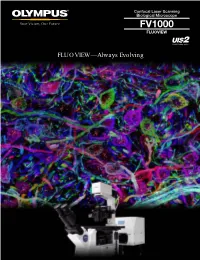
Fv1000 Fluoview
Confocal Laser Scanning Biological Microscope FV1000 FLUOVIEW FLUOVIEW—Always Evolving FLUOVIEW–—From Olympus is Open FLUOVIEW—More Advanced than Ever The Olympus FLUOVIEW FV1000 confocal laser scanning microscope delivers efficient and reliable performance together with the high resolution required for multi-dimensional observation of cell and tissue morphology, and precise molecular localization. The FV1000 incorporates the industry’s first dedicated laser light stimulation scanner to achieve simultaneous targeted laser stimulation and imaging for real-time visualization of rapid cell responses. The FV1000 also measures diffusion coefficients of intracellular molecules, quantifying molecular kinetics. Quite simply, the FLUOVIEW FV1000 represents a new plateau, bringing “imaging to analysis.” Olympus continues to drive forward the development of FLUOVIEW microscopes, using input from researchers to meet their evolving demands and bringing “imaging to analysis.” Quality Performance with Innovative Design FV10i 1 Imaging to Analysis ing up New Worlds From Imaging to Analysis FV1000 Advanced Deeper Imaging with High Resolution FV1000MPE 2 Advanced FLUOVIEW Systems Enhance the Power of Your Research Superb Optical Systems Set the Standard for Accuracy and Sensitivity. Two types of detectors deliver enhanced accuracy and sensitivity, and are paired with a new objective with low chromatic aberration, to deliver even better precision for colocalization analysis. These optical advances boost the overall system capabilities and raise performance to a new level. Imaging, Stimulation and Measurement— Advanced Analytical Methods for Quantification. Now equipped to measure the diffusion coefficients of intracellular molecules, for quantification of the dynamic interactions of molecules inside live cell. FLUOVIEW opens up new worlds of measurement. Evolving Systems Meet the Demands of Your Application. -

Two-Photon Excitation Fluorescence Microscopy
P1: FhN/ftt P2: FhN July 10, 2000 11:18 Annual Reviews AR106-15 Annu. Rev. Biomed. Eng. 2000. 02:399–429 Copyright c 2000 by Annual Reviews. All rights reserved TWO-PHOTON EXCITATION FLUORESCENCE MICROSCOPY PeterT.C.So1,ChenY.Dong1, Barry R. Masters2, and Keith M. Berland3 1Department of Mechanical Engineering, Massachusetts Institute of Technology, Cambridge, Massachusetts 02139; e-mail: [email protected] 2Department of Ophthalmology, University of Bern, Bern, Switzerland 3Department of Physics, Emory University, Atlanta, Georgia 30322 Key Words multiphoton, fluorescence spectroscopy, single molecule, functional imaging, tissue imaging ■ Abstract Two-photon fluorescence microscopy is one of the most important re- cent inventions in biological imaging. This technology enables noninvasive study of biological specimens in three dimensions with submicrometer resolution. Two-photon excitation of fluorophores results from the simultaneous absorption of two photons. This excitation process has a number of unique advantages, such as reduced specimen photodamage and enhanced penetration depth. It also produces higher-contrast im- ages and is a novel method to trigger localized photochemical reactions. Two-photon microscopy continues to find an increasing number of applications in biology and medicine. CONTENTS INTRODUCTION ................................................ 400 HISTORICAL REVIEW OF TWO-PHOTON MICROSCOPY TECHNOLOGY ...401 BASIC PRINCIPLES OF TWO-PHOTON MICROSCOPY ..................402 Physical Basis for Two-Photon Excitation ............................ -

SPARQ-Ed Risk Assessment Sheet : Immunofluorescence
SPARQ-ed Risk Assessment Sheet : Immunofluorescence Hazard Analyse / Evaluate Risk Overall Risk Category Description of Risk Source Current Controls Event Category Consequences Exposure Probability (see explanation on last page) Exposure PPE worn (gloves, closed Prob footwear, and safety Chemical exposure - VR R U O F C Exposure to Chemical Agents : Risk of Unusual : goggles provided). Use body contact, spills Unusual but eye and skin exposure to chemicals Immuno- AC Low Low Low Low Mod Subs of potentially harmful and splash and Other Contact Minor : General Possible: Less likely (such as fixation / permeabilisation fluorescence QP Low Low Low Low Low Mod Chemical chemicals/substances in inhalation (eg. 4% with Chemical or chemicals used would to occur with the buffer containing 4% staining not a UP Low Low Low Low Low Low fume cupboard/safety paraformaldehyde is Substance be irritants only. control measures but paraformaldehyde) throughout the common cabinet. MSDS available. irritant to eyes and not impossible. RP Low Low Low Low Low Low staining procedure. procedure. Well documented skin). C Low Low Low Low Low Low procedures. PI Low Low Low Low Low Low Personal precaution Exposure procedures to be Prob followed in case of VR R U O F C Sharps : Immunoflourescence is injury. PPE worn Sharps injury (cuts or Unusual : AC Low Low Low Low Mod Subs generally performed with sections on (wearing gloves, closed slicing of fingers) Immuno- Conceivable : Sharps slides or with cells on coverslips. shoes, lab coat and from broken Hitting Objects Minor : May require fluorescence injury is likely to QP Low Low Low Low Low Mod Immunofluorescence staining of cells Mechanical safety goggles are coverslips or pointed with Part of first aid for minor cuts. -

Etude Des Techniques De Super-Résolution Latérale En Nanoscopie Et Développement D’Un Système Interférométrique Nano-3D Audrey Leong-Hoï
Etude des techniques de super-résolution latérale en nanoscopie et développement d’un système interférométrique nano-3D Audrey Leong-Hoï To cite this version: Audrey Leong-Hoï. Etude des techniques de super-résolution latérale en nanoscopie et développement d’un système interférométrique nano-3D. Micro et nanotechnologies/Microélectronique. Université de Strasbourg, 2016. Français. NNT : 2016STRAD048. tel-02003485 HAL Id: tel-02003485 https://tel.archives-ouvertes.fr/tel-02003485 Submitted on 1 Feb 2019 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. UNIVERSITÉ DE STRASBOURG ÉCOLE DOCTORALE MATHEMATIQUES, SCIENCES DE L'INFORMATION ET DE L'INGENIEUR (MSII) – ED 269 LABORATOIRE DES SCIENCES DE L'INGENIEUR, DE L'INFORMATIQUE ET DE L'IMAGERIE (ICUBE UMR 7357) THÈSE présentée par : Audrey LEONG-HOI soutenue le : 2 DÉCEMBRE 2016 pour obtenir le grade de : Docteur de l’université de Strasbourg Discipline / Spécialité : Electronique, microélectronique, photonique Étude des techniques de super-résolution latérale en nanoscopie et développement d'un système interférométrique nano-3D THÈSE dirigée par : Dr. MONTGOMERY Paul Directeur de recherche, CNRS, ICube (Strasbourg) Pr. SERIO Bruno Professeur des Universités, Université Paris Ouest, LEME (Paris) RAPPORTEURS : Dr. GORECKI Christophe Directeur de recherche, CNRS, FEMTO-ST (Besançon) Pr. -

Label-Free Multiphoton Microscopy: Much More Than Fancy Images
International Journal of Molecular Sciences Review Label-Free Multiphoton Microscopy: Much More than Fancy Images Giulia Borile 1,2,*,†, Deborah Sandrin 2,3,†, Andrea Filippi 2, Kurt I. Anderson 4 and Filippo Romanato 1,2,3 1 Laboratory of Optics and Bioimaging, Institute of Pediatric Research Città della Speranza, 35127 Padua, Italy; fi[email protected] 2 Department of Physics and Astronomy “G. Galilei”, University of Padua, 35131 Padua, Italy; [email protected] (D.S.); andrea.fi[email protected] (A.F.) 3 L.I.F.E.L.A.B. Program, Consorzio per la Ricerca Sanitaria (CORIS), Veneto Region, 35128 Padua, Italy 4 Crick Advanced Light Microscopy Facility (CALM), The Francis Crick Institute, London NW1 1AT, UK; [email protected] * Correspondence: [email protected] † These authors contributed equally. Abstract: Multiphoton microscopy has recently passed the milestone of its first 30 years of activity in biomedical research. The growing interest around this approach has led to a variety of applications from basic research to clinical practice. Moreover, this technique offers the advantage of label-free multiphoton imaging to analyze samples without staining processes and the need for a dedicated system. Here, we review the state of the art of label-free techniques; then, we focus on two-photon autofluorescence as well as second and third harmonic generation, describing physical and technical characteristics. We summarize some successful applications to a plethora of biomedical research fields and samples, underlying the versatility of this technique. A paragraph is dedicated to an overview of sample preparation, which is a crucial step in every microscopy experiment. -

Label-Free Non-Linear Multimodal Optical Microscopy-Basics, Development, and Applications
Label-Free Non-linear Multimodal Optical Microscopy-Basics, Development, and Applications Nirmal Mazumder, Naveen Balla, Guan-Yu Zhuo, Yury Kistenev, Rajesh Kumar, Fu-Jen Kao, Sophie Brasselet, Viktor Nikolaev, Natalya Krivova To cite this version: Nirmal Mazumder, Naveen Balla, Guan-Yu Zhuo, Yury Kistenev, Rajesh Kumar, et al.. Label- Free Non-linear Multimodal Optical Microscopy-Basics, Development, and Applications. Frontiers in Physics, Frontiers, 2019, 7, 10.3389/fphy.2019.00170. hal-02410671 HAL Id: hal-02410671 https://hal.archives-ouvertes.fr/hal-02410671 Submitted on 13 Dec 2019 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. REVIEW published: 31 October 2019 doi: 10.3389/fphy.2019.00170 Label-Free Non-linear Multimodal Optical Microscopy—Basics, Development, and Applications Nirmal Mazumder 1*, Naveen K. Balla 2, Guan-Yu Zhuo 3,4, Yury V. Kistenev 5,6, Rajesh Kumar 7, Fu-Jen Kao 8, Sophie Brasselet 9, Viktor V. Nikolaev 5,10 and Natalya A. Krivova 11 1 Department of Biophysics, Manipal School of Life Sciences, Manipal Academy of Higher -

Super-Resolution Microscopy 1
Semrock White Paper Series: Super-resolution Microscopy 1. Introduction Fluorescence microscopy has revolutionized the study of biological samples. Ever since the invention of fluorescence microscopy towards the beginning of the 20th century, significant technological advances have enabled elucidation of biological phenomenon at cellular, sub- cellular and even at molecular levels. However, the latest incarnation of the modern fluorescence microscope has led to a paradigm shift. This wave is about breaking the diffraction limit first proposed in 1873 by Ernst Abbe. The implications of this development are profound. This new technology, called super-resolution microscopy, allows for the visualization of cellular samples with a resolution similar to that of an electron microscope, yet it retains the advantages of an optical fluorescence microscope. This means it is possible to uniquely visualize desired molecular species in a cellular environment, even in three dimensions and now in live cells – all at a scale comparable to the spatial dimensions of the molecules under investigation. This article provides an overview of some of the key recent developments in super-resolution microscopy. 2. The problem So what is wrong with a conventional fluorescence microscope? To understand the answer to this question, consider a Green Fluorescence Protein (GFP) molecule that is about 3 nm in diameter and about 4 nm long. When a single GFP molecule is imaged using a 100X objective the size of the image should be 0.3 to 0.4 microns (100 times the size of the object). Yet the smallest spot that can be seen at the camera is about 25 microns. Which corresponds to an object size of about 250 nm. -

How Single-Molecule Localization Microscopy Expandedour
International Journal of Molecular Sciences Review How Single-Molecule Localization Microscopy Expanded Our Mechanistic Understanding of RNA Polymerase II Transcription Peter Hoboth 1,2 , OndˇrejŠebesta 2 and Pavel Hozák 1,3,* 1 Department of Biology of the Cell Nucleus, Institute of Molecular Genetics of the Czech Academy of Sciences, Vídeˇnská 1083, 142 20 Prague, Czech Republic; [email protected] 2 Faculty of Science, Charles University, Albertov 6, 128 00 Prague, Czech Republic; [email protected] 3 Microscopy Centre, Institute of Molecular Genetics of the Czech Academy of Sciences, Vídeˇnská 1083, 142 20 Prague, Czech Republic * Correspondence: [email protected] Abstract: Classical models of gene expression were built using genetics and biochemistry. Although these approaches are powerful, they have very limited consideration of the spatial and temporal organization of gene expression. Although the spatial organization and dynamics of RNA polymerase II (RNAPII) transcription machinery have fundamental functional consequences for gene expression, its detailed studies have been abrogated by the limits of classical light microscopy for a long time. The advent of super-resolution microscopy (SRM) techniques allowed for the visualization of the RNAPII transcription machinery with nanometer resolution and millisecond precision. In this review, we summarize the recent methodological advances in SRM, focus on its application for studies of the nanoscale organization in space and time of RNAPII transcription, and discuss its consequences for the mechanistic understanding of gene expression. Citation: Hoboth, P.; Šebesta, O.; Hozák, P. How Single-Molecule Keywords: cell nucleus; gene expression; transcription foci; transcription factors; super-resolution Localization Microscopy Expanded microscopy; structured illumination; stimulated emission depletion; stochastic optical reconstruc- Our Mechanistic Understanding of tion; photoactivation RNA Polymerase II Transcription. -

Super-Resolved Fluorescence Microscopy
8 OCTOBER 2014 Scientifc Background on the Nobel Prize in Chemistry 2014 SUPER-RESOLVED FLUORESCENCE MICROSCOPY THE ROYAL SWEDISH ACADEMY OF SCIENCES has as its aim to promote the sciences and strengthen their infuence in society. BOX 50005 (LILLA FRESCATIVÄGEN 4 A), SE-104 05 STOCKHOLM, SWEDEN Nobel Prize® and the Nobel Prize® medal design mark TEL +46 8 673 95 00, [email protected] HTTP://KVA.SE are registrated trademarks of the Nobel Foundation Super-resolved fluorescence microscopy The Royal Swedish Academy of Sciences has decided to award Eric Betzig, Stefan W. Hell and W. E. Moerner the Nobel Prize in Chemistry 2014 for the development of super-resolution fluorescence microscopy. 1. Abbe’s diffraction limit Towards the end of the nineteenth century Ernst Abbe (Abbe, 1873) and Lord Rayleigh (Rayleigh, 1896) formulated what is commonly known as the “diffraction limit” for microscopy. Roughly speaking, this limit states that it is impossible to resolve two elements of a structure which are closer to each other than about half the wave length (λ) in the lateral (x,y) plane and even further apart in the longitudinal plane (z). In other words, the minimal distances (δxmin, δymin) that, according to Abbe’s criterion, can be resolved in the lateral plane are approximated by xymin, min / 2 Another consequence of the same diffraction limitation is that it is not possible to focus a laser beam to a spot of smaller dimension than about λ/2 (Fig. 1). Figure 1: (Huang et al., 2010, Cell, 143, 1047 – 57). A: (left) Focused laser beam, (middle) structure, (right) resolved (A) and not resolved (B) structural features. -
Basic Fluorescence Microscopy and Sample Preparation
Basic Fluorescence Microscopy and Sample Preparation Eva Wegel [email protected] Visible Light 390 – 700 nm visible to the human eye White light is split into its components through a prism Reason: different λ refract at different angles What is fluorescence? George Gabriel Stokes (1819-1903) Jablonski diagram Stokes Shift Alexa Fluor 488 Photoluminescence: Fluorescence - spontaneous emission of light during transition of the system from its lowest vibrational -9 -6 energy level of an excited singlet state S1 back to the ground state S0 (10 to 10 s) Phosphorescence – a non-radiative transition into an isoenergetic vibrational level of a triplet state T1 , which lasts for 10-3 to 1000 s before it decays to the ground state IC: internal conversion ISC: intersystem crossing Photobleaching and Phototoxicity Photobleaching: photochemical destruction of the fluorophore In an excited triplet state, fluorophores may interact with another molecule to produce irreversible covalent modifications Phototoxicity: illumination of a fluorophore causes damage to the cell expressing it, eventually leading to cell death Common situation: the excited dye molecule passes its excess energy on to O2, creating reactive oxygen species (ROS): - ROS reacts with dye dye bleaches - ROS diffuses away and reacts with other dyes or cell components Solution: - Reduce the intensity of the excitation light and frequency of illumination - Close down the field aperture in order to restrict the illuminated area Basic principle of an (Inverted) Fluorescence Microscope