The Convoluted Evolutionary History of the Capped-Golden Langur Lineage 2 (Cercopithecidae: Colobinae) – Concatenation Versus Coalescent Analyses
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Body Measurements for the Monkeys of Bioko Island, Equatorial Guinea
Primate Conservation 2009 (24): 99–105 Body Measurements for the Monkeys of Bioko Island, Equatorial Guinea Thomas M. Butynski¹,², Yvonne A. de Jong² and Gail W. Hearn¹ ¹Bioko Biodiversity Protection Program, Drexel University, Philadelphia, PA, USA ²Eastern Africa Primate Diversity and Conservation Program, Nanyuki, Kenya Abstract: Bioko Island, Equatorial Guinea, has a rich (eight genera, 11 species), unique (seven endemic subspecies), and threat- ened (five species) primate fauna, but the taxonomic status of most forms is not clear. This uncertainty is a serious problem for the setting of priorities for the conservation of Bioko’s (and the region’s) primates. Some of the questions related to the taxonomic status of Bioko’s primates can be resolved through the statistical comparison of data on their body measurements with those of their counterparts on the African mainland. Data for such comparisons are, however, lacking. This note presents the first large set of body measurement data for each of the seven species of monkeys endemic to Bioko; means, ranges, standard deviations and sample sizes for seven body measurements. These 49 data sets derive from 544 fresh adult specimens (235 adult males and 309 adult females) collected by shotgun hunters for sale in the bushmeat market in Malabo. Key Words: Bioko Island, body measurements, conservation, monkeys, morphology, taxonomy Introduction gordonorum), and surprisingly few such data exist even for some of the more widespread species (for example, Allen’s Comparing external body measurements for adult indi- swamp monkey Allenopithecus nigroviridis, northern tala- viduals from different sites has long been used as a tool for poin monkey Miopithecus ogouensis, and grivet Chlorocebus describing populations, subspecies, and species of animals aethiops). -

Habitat Utilization and Feeding Biology of Himalayan Grey Langur
动 物 学 研 究 2010,Apr. 31(2):177−188 CN 53-1040/Q ISSN 0254-5853 Zoological Research DOI:10.3724/SP.J.1141.2010.02177 Habitat Utilization and Feeding Biology of Himalayan Grey Langur (Semnopithecus entellus ajex) in Machiara National Park, Azad Jammu and Kashmir, Pakistan Riaz Aziz Minhas1,*, Khawaja Basharat Ahmed2, Muhammad Siddique Awan2, Naeem Iftikhar Dar3 (1. World Wide Fund for Nature-Pakistan (WWF-Pakistan) AJK Office Muzaffarabad, Azad Jammu and Kashmir 13100, Pakistan; 2. Department ofZoology, University of Azad Jammu and Kashmir Muzaffarabad, Azad Jammu and Kashmir 13100, Pakistan; 3. Department of Wildlife and Fisheries, Government of Azad Jammu and Kashmir, Muzaffarabad, Azad Jammu and Kashmir 13100, Pakistan) Abstract: Habitat utilization and feeding biology of Himalayan Grey Langur (Semnopithecus entellus ajex) were studied from April, 2006 to April, 2007 in Machiara National Park, Azad Jammu and Kashmir, Pakistan. The results showed that in the winter season the most preferred habitat of the langurs was the moist temperate coniferous forests interspersed with deciduous trees, while in the summer season they preferred to migrate into the subalpine scrub forests at higher altitudes. Langurs were folivorous in feeding habit, recorded as consuming more than 49 plant species (27 in summer and 22 in winter) in the study area. The mature leaves (36.12%) were preferred over the young leaves (27.27%) while other food components comprised of fruits (17.00%), roots (9.45%), barks (6.69%), flowers (2.19%) and stems (1.28%) of various plant species. Key words: Himalayan Grey Langur; Habitat; Food biology; Machiara National Park 喜马拉雅灰叶猴栖息地利用和食性生物学 Riaz Aziz Minhas1,*, Khawaja Basharat Ahmed2, Muhammad Siddique Awan2, Naeem Iftikhar Dar3 (1. -

Controlled Animals
Environment and Sustainable Resource Development Fish and Wildlife Policy Division Controlled Animals Wildlife Regulation, Schedule 5, Part 1-4: Controlled Animals Subject to the Wildlife Act, a person must not be in possession of a wildlife or controlled animal unless authorized by a permit to do so, the animal was lawfully acquired, was lawfully exported from a jurisdiction outside of Alberta and was lawfully imported into Alberta. NOTES: 1 Animals listed in this Schedule, as a general rule, are described in the left hand column by reference to common or descriptive names and in the right hand column by reference to scientific names. But, in the event of any conflict as to the kind of animals that are listed, a scientific name in the right hand column prevails over the corresponding common or descriptive name in the left hand column. 2 Also included in this Schedule is any animal that is the hybrid offspring resulting from the crossing, whether before or after the commencement of this Schedule, of 2 animals at least one of which is or was an animal of a kind that is a controlled animal by virtue of this Schedule. 3 This Schedule excludes all wildlife animals, and therefore if a wildlife animal would, but for this Note, be included in this Schedule, it is hereby excluded from being a controlled animal. Part 1 Mammals (Class Mammalia) 1. AMERICAN OPOSSUMS (Family Didelphidae) Virginia Opossum Didelphis virginiana 2. SHREWS (Family Soricidae) Long-tailed Shrews Genus Sorex Arboreal Brown-toothed Shrew Episoriculus macrurus North American Least Shrew Cryptotis parva Old World Water Shrews Genus Neomys Ussuri White-toothed Shrew Crocidura lasiura Greater White-toothed Shrew Crocidura russula Siberian Shrew Crocidura sibirica Piebald Shrew Diplomesodon pulchellum 3. -

The Role of Exposure in Conservation
Behavioral Application in Wildlife Photography: Developing a Foundation in Ecological and Behavioral Characteristics of the Zanzibar Red Colobus Monkey (Procolobus kirkii) as it Applies to the Development Exhibition Photography Matthew Jorgensen April 29, 2009 SIT: Zanzibar – Coastal Ecology and Natural Resource Management Spring 2009 Advisor: Kim Howell – UDSM Academic Director: Helen Peeks Table of Contents Acknowledgements – 3 Abstract – 4 Introduction – 4-15 • 4 - The Role of Exposure in Conservation • 5 - The Zanzibar Red Colobus (Piliocolobus kirkii) as a Conservation Symbol • 6 - Colobine Physiology and Natural History • 8 - Colobine Behavior • 8 - Physical Display (Visual Communication) • 11 - Vocal Communication • 13 - Olfactory and Tactile Communication • 14 - The Importance of Behavioral Knowledge Study Area - 15 Methodology - 15 Results - 16 Discussion – 17-30 • 17 - Success of the Exhibition • 18 - Individual Image Assessment • 28 - Final Exhibition Assessment • 29 - Behavioral Foundation and Photography Conclusion - 30 Evaluation - 31 Bibliography - 32 Appendices - 33 2 To all those who helped me along the way, I am forever in your debt. To Helen Peeks and Said Hamad Omar for a semester of advice, and for trying to make my dreams possible (despite the insurmountable odds). Ali Ali Mwinyi, for making my planning at Jozani as simple as possible, I thank you. I would like to thank Bi Ashura, for getting me settled at Jozani and ensuring my comfort during studies. Finally, I am thankful to the rangers and staff of Jozani for welcoming me into the park, for their encouragement and support of my project. To Kim Howell, for agreeing to support a project outside his area of expertise, I am eternally grateful. -
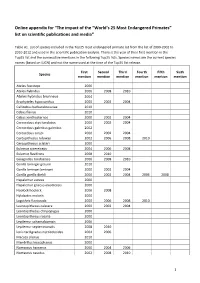
Online Appendix for “The Impact of the “World's 25 Most Endangered
Online appendix for “The impact of the “World’s 25 Most Endangered Primates” list on scientific publications and media” Table A1. List of species included in the Top25 most endangered primate list from the list of 2000-2002 to 2010-2012 and used in the scientific publication analysis. There is the year of their first mention in the Top25 list and the consecutive mentions in the following Top25 lists. Species names are the current species names (based on IUCN) and not the name used at the time of the Top25 list release. First Second Third Fourth Fifth Sixth Species mention mention mention mention mention mention Ateles fusciceps 2006 Ateles hybridus 2006 2008 2010 Ateles hybridus brunneus 2004 Brachyteles hypoxanthus 2000 2002 2004 Callicebus barbarabrownae 2010 Cebus flavius 2010 Cebus xanthosternos 2000 2002 2004 Cercocebus atys lunulatus 2000 2002 2004 Cercocebus galeritus galeritus 2002 Cercocebus sanjei 2000 2002 2004 Cercopithecus roloway 2002 2006 2008 2010 Cercopithecus sclateri 2000 Eulemur cinereiceps 2004 2006 2008 Eulemur flavifrons 2008 2010 Galagoides rondoensis 2006 2008 2010 Gorilla beringei graueri 2010 Gorilla beringei beringei 2000 2002 2004 Gorilla gorilla diehli 2000 2002 2004 2006 2008 Hapalemur aureus 2000 Hapalemur griseus alaotrensis 2000 Hoolock hoolock 2006 2008 Hylobates moloch 2000 Lagothrix flavicauda 2000 2006 2008 2010 Leontopithecus caissara 2000 2002 2004 Leontopithecus chrysopygus 2000 Leontopithecus rosalia 2000 Lepilemur sahamalazensis 2006 Lepilemur septentrionalis 2008 2010 Loris tardigradus nycticeboides -

Mandrillus Leucophaeus Poensis)
Ecology and Behavior of the Bioko Island Drill (Mandrillus leucophaeus poensis) A Thesis Submitted to the Faculty of Drexel University by Jacob Robert Owens in partial fulfillment of the requirements for the degree of Doctor of Philosophy December 2013 i © Copyright 2013 Jacob Robert Owens. All Rights Reserved ii Dedications To my wife, Jen. iii Acknowledgments The research presented herein was made possible by the financial support provided by Primate Conservation Inc., ExxonMobil Foundation, Mobil Equatorial Guinea, Inc., Margo Marsh Biodiversity Fund, and the Los Angeles Zoo. I would also like to express my gratitude to Dr. Teck-Kah Lim and the Drexel University Office of Graduate Studies for the Dissertation Fellowship and the invaluable time it provided me during the writing process. I thank the Government of Equatorial Guinea, the Ministry of Fisheries and the Environment, Ministry of Information, Press, and Radio, and the Ministry of Culture and Tourism for the opportunity to work and live in one of the most beautiful and unique places in the world. I am grateful to the faculty and staff of the National University of Equatorial Guinea who helped me navigate the geographic and bureaucratic landscape of Bioko Island. I would especially like to thank Jose Manuel Esara Echube, Claudio Posa Bohome, Maximilliano Fero Meñe, Eusebio Ondo Nguema, and Mariano Obama Bibang. The journey to my Ph.D. has been considerably more taxing than I expected, and I would not have been able to complete it without the assistance of an expansive list of people. I would like to thank all of you who have helped me through this process, many of whom I lack the space to do so specifically here. -

Common Chimpanzees (Pan Troglodytes), Moun
KroeberAnthropological Society Papers, Nos. 71-72, 1990 Colobine Socioecology and Female-bonded Models of Primate Social Structure Craig B. Stanford Ecological models ofprimate social systems have been used extensively to explain the variationsfound in social organization among living primates and to accountforprimate sociality itself. Recent attempts to characterize primate social systems as either 'female-bonded" or "non-female-bonded" establish a typology that does notfully consider the variation inpatterns ofsex-biased dispersal seen in the Primate order. Thispaper uses as its example the Old World monkey subfamily Colobinae to show that ecologi- cal models are basedprimarily onfrugivorous, territorialpinmates. The models are inadequate to explain patterns ofintergroup competition and should not be usedfor setting general rulesforprimate societies. Apreliminary alternative view ofsex-biased dispersal that is basedonfrequency dependence is offered. INTRODUCTION bonded according to each species' typical pattern Trivers (1972) and Emlen and Oring (1977) of sex-biased dispersal. Males are assumed to outlined the hypothesis that females and males have a greater lifetime reproductive potential than have been selected to invest their lifetime energies females, and in most species they invest less in differently: females in maintaining access to offspring than do females (Trivers 1972). Since maintenance and growth resources, in order to food intake for an individual female primate is invest most in their offspring; and males in a maximized by feeding singly, the evolution of reproductive strategy that maximizes access to fe- primate sociality suggests that there is some bene- males, investing relatively little in offspring. In fit accruing to females who forage as a group. short, females should compete for food, and Wrangham (1980) considered this benefit, which males should compete for females. -

Semnopithecus Entellus) in the ARAVALLI HILLS of RAJASTHAN, INDIA
Contents TIGERPAPER Population Ecology of Hanuman Langurs in the Aravalli Hills of Rajasthan, India.................................................................. 1 Status and Conservation of Gangetic Dolphin in the Karnali River, Nepal........................................................................... 8 Population Estimation of Tigers in Maharashtra............................. 11 Use of Capped Langur Habitat Suitability Analysis in Planning And Management of Upland Protected Areas in Bangladesh......... 13 Eco-Friendly Management of Rose-Ringed Parakeet in Winter Maize Crop................................................................. 23 The Wildlife Value: Example from West Papua, Indonesia.............. 27 The Butterfly Fauna of Visakhapatnam in South India.................... 30 FOREST NEWS What Were the Most Significant Developments in the Forestry Sector in Asia-Pacific in 2002?................................................. 1 Update on the Search for Excellence in Forest Management in Asia-Pacific.......................................................................... 6 Seeking New Highs for Mountain Area Development..................... 8 Adaptive Collaborative Management of Community Forests: An Option for Asia?................................................................ 8 Staff Movement........................................................................ 9 What’s the Role of Forests in Sustainable Water Management? 10 New RAP Forestry Publications.................................................. 12 -

Gastrointestinal Parasites of the Colobus Monkeys of Uganda
J. Parasitol., 91(3), 2005, pp. 569±573 q American Society of Parasitologists 2005 GASTROINTESTINAL PARASITES OF THE COLOBUS MONKEYS OF UGANDA Thomas R. Gillespie*², Ellis C. Greiner³, and Colin A. Chapman²§ Department of Zoology, University of Florida, Gainesville, Florida 32611. e-mail: [email protected] ABSTRACT: From August 1997 to July 2003, we collected 2,103 fecal samples from free-ranging individuals of the 3 colobus monkey species of UgandaÐthe endangered red colobus (Piliocolobus tephrosceles), the eastern black-and-white colobus (Co- lobus guereza), and the Angolan black-and-white colobus (C. angolensis)Ðto identify and determine the prevalence of gastro- intestinal parasites. Helminth eggs, larvae, and protozoan cysts were isolated by sodium nitrate ¯otation and fecal sedimentation. Coprocultures facilitated identi®cation of helminths. Seven nematodes (Strongyloides fulleborni, S. stercoralis, Oesophagostomum sp., an unidenti®ed strongyle, Trichuris sp., Ascaris sp., and Colobenterobius sp.), 1 cestode (Bertiella sp.), 1 trematode (Dicro- coeliidae), and 3 protozoans (Entamoeba coli, E. histolytica, and Giardia lamblia) were detected. Seasonal patterns of infection were not apparent for any parasite species infecting colobus monkeys. Prevalence of S. fulleborni was higher in adult male compared to adult female red colobus, but prevalence did not differ for any other shared parasite species between age and sex classes. Colobinae is a large subfamily of leaf-eating, Old World 19 from Angolan black-and-white colobus. Red colobus are sexually monkeys represented in Africa by species of 3 genera, Colobus, dimorphic, with males averaging 10.5 kg and females 7.0 kg (Oates et al., 1994); they display a multimale±multifemale social structure and Procolobus, and Piliocolobus (Grubb et al., 2002). -

Bioko Red Colobus Piliocolobus Pennantii Pennantii (Waterhouse, 1838) Bioko Island, Equatorial Guinea (2004, 2006, 2010, 2012)
Bioko Red Colobus Piliocolobus pennantii pennantii (Waterhouse, 1838) Bioko Island, Equatorial Guinea (2004, 2006, 2010, 2012) Drew T. Cronin, Gail W. Hearn & John F. Oates Bioko red colobus (Piliocolobus pennantii pennantii) (Illustration: Stephen D. Nash) Pennant’s red colobus monkey Piliocolobus pennantii is P. p. pennantii is threatened by bushmeat hunting, presently regarded by the IUCN Red List as comprising most notably since the early 1980s when a commercial three subspecies: P. pennantii pennantii of Bioko, P. p. bushmeat market appeared in the town of Malabo epieni of the Niger Delta, and P. p. bouvieri of the Congo (Butynski and Koster 1994). Following the discovery Republic. Some accounts give full species status to of offshore oil in 1996, and the subsequent expansion all three of these monkeys (Groves 2007; Oates 2011; of Equatorial Guinea’s economy, rising urban demand Groves and Ting 2013). P. p. pennantii is currently led to increased numbers of primate carcasses in the classified as Endangered (Oates and Struhsaker 2008). bushmeat market (Morra et al. 2009; Cronin 2013). In November 2007, a primate hunting ban was enacted Piliocolobus pennantii pennantii may once have occurred on Bioko, but it lacked any realistic enforcement and over most of Bioko, but it is now probably limited to an contributed to a spike in the numbers of monkeys in the area of less than 300 km² within the Gran Caldera and market. Between October 1997 and September 2010, a 510 km² range in the Southern Highlands Scientific a total of 1,754 P. p. pennantii were observed for sale Reserve (GCSH) (Cronin et al. -

Phayre's Langur in Satchari National Park, Bangladesh
10 Asian Primates Journal 9(1), 2021 STATUS OF PHAYRE’S LANGUR Trachypithecus phayrei IN SATCHARI NATIONAL PARK, BANGLADESH Hassan Al-Razi1 and Habibon Naher2* Department of Zoology, Jagannath University, 9-11 Chittaranjan Avenue, Dhaka-1100, Bangladesh.1Email: chayan1999@ yahoo.com, 2Email: [email protected]. *Corresponding author ABSTRACT We studied the population status of Phayre’s Langur in Satchari National Park, Bangladesh, and threats to this population, from January to December 2016. We recorded 23 individuals in three groups. Group size ranged from four to 12 (mean 7.7±4.0) individuals; all groups contained a single adult male, 1–4 females and 2–7 immature individuals (subadults, juveniles and infants). Habitat encroachment for expansion of lemon orchards by the Tipra ethnic community and habitat degradation due to logging and firewood collection are the main threats to the primates. Road mortality, electrocution and tourist activities were additional causes of stress and mortality. Participatory work and awareness programmes with the Tipra community or generation of alternative income sources may reduce the dependency of local people on forest resources. Strict implementation of the rules and regulations of the Bangladesh Wildlife (Security and Conservation) Act 2012 can limit habitat encroachment and illegal logging, which should help in the conservation of this species. Key Words: Group composition, habitat encroachment, Satchari National Park. INTRODUCTION Phayre’s Langur (Phayre’s Leaf Monkey, Spectacled (1986) recorded 15 Phayre’s Langur groups comprising Langur) Trachypithecus phayrei (Blyth) occurs in 205 individuals in the north-east and south-east of Bangladesh, China, India and Myanmar (Bleisch et al., Bangladesh. -

Fossil Primates
AccessScience from McGraw-Hill Education Page 1 of 16 www.accessscience.com Fossil primates Contributed by: Eric Delson Publication year: 2014 Extinct members of the order of mammals to which humans belong. All current classifications divide the living primates into two major groups (suborders): the Strepsirhini or “lower” primates (lemurs, lorises, and bushbabies) and the Haplorhini or “higher” primates [tarsiers and anthropoids (New and Old World monkeys, greater and lesser apes, and humans)]. Some fossil groups (omomyiforms and adapiforms) can be placed with or near these two extant groupings; however, there is contention whether the Plesiadapiformes represent the earliest relatives of primates and are best placed within the order (as here) or outside it. See also: FOSSIL; MAMMALIA; PHYLOGENY; PHYSICAL ANTHROPOLOGY; PRIMATES. Vast evidence suggests that the order Primates is a monophyletic group, that is, the primates have a common genetic origin. Although several peculiarities of the primate bauplan (body plan) appear to be inherited from an inferred common ancestor, it seems that the order as a whole is characterized by showing a variety of parallel adaptations in different groups to a predominantly arboreal lifestyle, including anatomical and behavioral complexes related to improved grasping and manipulative capacities, a variety of locomotor styles, and enlargement of the higher centers of the brain. Among the extant primates, the lower primates more closely resemble forms that evolved relatively early in the history of the order, whereas the higher primates represent a group that evolved more recently (Fig. 1). A classification of the primates, as accepted here, appears above. Early primates The earliest primates are placed in their own semiorder, Plesiadapiformes (as contrasted with the semiorder Euprimates for all living forms), because they have no direct evolutionary links with, and bear few adaptive resemblances to, any group of living primates.