Journal of Down Syndrome & Chromosome Abnormalities
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Large-Scale Analysis of Genome and Transcriptome Alterations in Multiple Tumors Unveils Novel Cancer-Relevant Splicing Networks
Downloaded from genome.cshlp.org on September 28, 2021 - Published by Cold Spring Harbor Laboratory Press Research Large-scale analysis of genome and transcriptome alterations in multiple tumors unveils novel cancer-relevant splicing networks Endre Sebestyén,1,5 Babita Singh,1,5 Belén Miñana,1,2 Amadís Pagès,1 Francesca Mateo,3 Miguel Angel Pujana,3 Juan Valcárcel,1,2,4 and Eduardo Eyras1,4 1Universitat Pompeu Fabra, E08003 Barcelona, Spain; 2Centre for Genomic Regulation, E08003 Barcelona, Spain; 3Program Against Cancer Therapeutic Resistance (ProCURE), Catalan Institute of Oncology (ICO), Bellvitge Institute for Biomedical Research (IDIBELL), E08908 L’Hospitalet del Llobregat, Spain; 4Catalan Institution for Research and Advanced Studies, E08010 Barcelona, Spain Alternative splicing is regulated by multiple RNA-binding proteins and influences the expression of most eukaryotic genes. However, the role of this process in human disease, and particularly in cancer, is only starting to be unveiled. We system- atically analyzed mutation, copy number, and gene expression patterns of 1348 RNA-binding protein (RBP) genes in 11 solid tumor types, together with alternative splicing changes in these tumors and the enrichment of binding motifs in the alter- natively spliced sequences. Our comprehensive study reveals widespread alterations in the expression of RBP genes, as well as novel mutations and copy number variations in association with multiple alternative splicing changes in cancer drivers and oncogenic pathways. Remarkably, the altered splicing patterns in several tumor types recapitulate those of undifferen- tiated cells. These patterns are predicted to be mainly controlled by MBNL1 and involve multiple cancer drivers, including the mitotic gene NUMA1. We show that NUMA1 alternative splicing induces enhanced cell proliferation and centrosome am- plification in nontumorigenic mammary epithelial cells. -

RBM11 Sirna (H): Sc-91387
SANTA CRUZ BIOTECHNOLOGY, INC. RBM11 siRNA (h): sc-91387 BACKGROUND PRODUCT Proteins containing RNA recognition motifs, including various hnRNP proteins, RBM11 siRNA (h) is a pool of 3 target-specific 19-25 nt siRNAs designed are implicated in the regulation of alternative splicing and protein components to knock down gene expression. Each vial contains 3.3 nmol of lyophilized of snRNPs. The RBM (RNA-binding motif) gene family encodes proteins with siRNA, sufficient for a 10 µM solution once resuspended using protocol an RNA binding motif that have been suggested to play a role in the modu- below. Suitable for 50-100 transfections. Also see RBM11 shRNA Plasmid (h): lation of apoptosis. RBM11 (RNA-binding protein 11) is a 281 amino acid sc-91387-SH and RBM11 shRNA (h) Lentiviral Particles: sc-91387-V as alter- nuclear protein that contains one RNA recognition motif. RBM11 exists as nate gene silencing products. two isoforms produced by alternative splicing and is expressed in testis, kid- For independent verification of RBM11 (h) gene silencing results, we also ney, spleen, brain, spinal cord and mammary gland. The gene that encodes provide the individual siRNA duplex components. Each is available as 3.3 nmol RBM11 maps to chromosome 21, the smallest of the human chromosomes. of lyophilized siRNA. These include: sc-91387A, sc-91387B and sc-91387C. Down syndrome, also known as trisomy 21, is the disease most commonly associated with chromosome 21. Alzheimer’s disease, Jervell and Lange- STORAGE AND RESUSPENSION Nielsen syndrome and amyotrophic lateral sclerosis are also associated with chromosome 21. Store lyophilized siRNA duplex at -20° C with desiccant. -

A Single-Cell Transcriptomic Landscape of Primate Arterial Aging
ARTICLE https://doi.org/10.1038/s41467-020-15997-0 OPEN A single-cell transcriptomic landscape of primate arterial aging Weiqi Zhang 1,2,3,4,5,13, Shu Zhang6,7,13, Pengze Yan3,8,13, Jie Ren7,9,13, Moshi Song3,5,8, Jingyi Li2,3,8, Jinghui Lei4, Huize Pan2,3, Si Wang3,5,8, Xibo Ma3,10, Shuai Ma2,3,8, Hongyu Li2,3, Fei Sun2,3, Haifeng Wan3,5,11, ✉ ✉ ✉ Wei Li 3,5,11, Piu Chan4, Qi Zhou3,5,11, Guang-Hui Liu 2,3,4,5,8 , Fuchou Tang 6,7,9,12 & Jing Qu 3,5,11 Our understanding of how aging affects the cellular and molecular components of the vas- 1234567890():,; culature and contributes to cardiovascular diseases is still limited. Here we report a single-cell transcriptomic survey of aortas and coronary arteries in young and old cynomolgus monkeys. Our data define the molecular signatures of specialized arteries and identify eight markers discriminating aortic and coronary vasculatures. Gene network analyses characterize tran- scriptional landmarks that regulate vascular senility and position FOXO3A, a longevity- associated transcription factor, as a master regulator gene that is downregulated in six subtypes of monkey vascular cells during aging. Targeted inactivation of FOXO3A in human vascular endothelial cells recapitulates the major phenotypic defects observed in aged monkey arteries, verifying FOXO3A loss as a key driver for arterial endothelial aging. Our study provides a critical resource for understanding the principles underlying primate arterial aging and contributes important clues to future treatment of age-associated vascular disorders. 1 CAS Key Laboratory of Genomic and Precision Medicine, Beijing Institute of Genomics, Chinese Academy of Sciences, Beijing 100101, China. -

Human Lectins, Their Carbohydrate Affinities and Where to Find Them
biomolecules Review Human Lectins, Their Carbohydrate Affinities and Where to Review HumanFind Them Lectins, Their Carbohydrate Affinities and Where to FindCláudia ThemD. Raposo 1,*, André B. Canelas 2 and M. Teresa Barros 1 1, 2 1 Cláudia D. Raposo * , Andr1 é LAQVB. Canelas‐Requimte,and Department M. Teresa of Chemistry, Barros NOVA School of Science and Technology, Universidade NOVA de Lisboa, 2829‐516 Caparica, Portugal; [email protected] 12 GlanbiaLAQV-Requimte,‐AgriChemWhey, Department Lisheen of Chemistry, Mine, Killoran, NOVA Moyne, School E41 of ScienceR622 Co. and Tipperary, Technology, Ireland; canelas‐ [email protected] NOVA de Lisboa, 2829-516 Caparica, Portugal; [email protected] 2* Correspondence:Glanbia-AgriChemWhey, [email protected]; Lisheen Mine, Tel.: Killoran, +351‐212948550 Moyne, E41 R622 Tipperary, Ireland; [email protected] * Correspondence: [email protected]; Tel.: +351-212948550 Abstract: Lectins are a class of proteins responsible for several biological roles such as cell‐cell in‐ Abstract:teractions,Lectins signaling are pathways, a class of and proteins several responsible innate immune for several responses biological against roles pathogens. such as Since cell-cell lec‐ interactions,tins are able signalingto bind to pathways, carbohydrates, and several they can innate be a immuneviable target responses for targeted against drug pathogens. delivery Since sys‐ lectinstems. In are fact, able several to bind lectins to carbohydrates, were approved they by canFood be and a viable Drug targetAdministration for targeted for drugthat purpose. delivery systems.Information In fact, about several specific lectins carbohydrate were approved recognition by Food by andlectin Drug receptors Administration was gathered for that herein, purpose. plus Informationthe specific organs about specific where those carbohydrate lectins can recognition be found by within lectin the receptors human was body. -

A Chromosome Level Genome of Astyanax Mexicanus Surface Fish for Comparing Population
bioRxiv preprint doi: https://doi.org/10.1101/2020.07.06.189654; this version posted July 6, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. 1 Title 2 A chromosome level genome of Astyanax mexicanus surface fish for comparing population- 3 specific genetic differences contributing to trait evolution. 4 5 Authors 6 Wesley C. Warren1, Tyler E. Boggs2, Richard Borowsky3, Brian M. Carlson4, Estephany 7 Ferrufino5, Joshua B. Gross2, LaDeana Hillier6, Zhilian Hu7, Alex C. Keene8, Alexander Kenzior9, 8 Johanna E. Kowalko5, Chad Tomlinson10, Milinn Kremitzki10, Madeleine E. Lemieux11, Tina 9 Graves-Lindsay10, Suzanne E. McGaugh12, Jeff T. Miller12, Mathilda Mommersteeg7, Rachel L. 10 Moran12, Robert Peuß9, Edward Rice1, Misty R. Riddle13, Itzel Sifuentes-Romero5, Bethany A. 11 Stanhope5,8, Clifford J. Tabin13, Sunishka Thakur5, Yamamoto Yoshiyuki14, Nicolas Rohner9,15 12 13 Authors for correspondence: Wesley C. Warren ([email protected]), Nicolas Rohner 14 ([email protected]) 15 16 Affiliation 17 1Department of Animal Sciences, Department of Surgery, Institute for Data Science and 18 Informatics, University of Missouri, Bond Life Sciences Center, Columbia, MO 19 2 Department of Biological Sciences, University of Cincinnati, Cincinnati, OH 20 3 Department of Biology, New York University, New York, NY 21 4 Department of Biology, The College of Wooster, Wooster, OH 22 5 Harriet L. Wilkes Honors College, Florida Atlantic University, Jupiter FL 23 6 Department of Genome Sciences, University of Washington, Seattle, WA 1 bioRxiv preprint doi: https://doi.org/10.1101/2020.07.06.189654; this version posted July 6, 2020. -

The Expression of Cold-Inducible RNA-Binding Protein Mrna in Sow Genital Tract Is Modulated by Natural Mating, but Not by Seminal Plasma
International Journal of Molecular Sciences Article The Expression of Cold-Inducible RNA-Binding Protein mRNA in Sow Genital Tract Is Modulated by Natural Mating, but not by Seminal Plasma Jaume Gardela 1,2 , Mateo Ruiz-Conca 1,2 , Cristina A. Martinez 1 , Dominic Wright 3, Manel López-Béjar 2,4, Heriberto Rodriguez-Martinez 1 and Manuel Alvarez-Rodriguez 1,2,* 1 Department of Biomedical and Clinical Sciences (BKV), Division of Children’s and Women Health (BKH), Obstetrics and Gynaecology, Linköping University, 58185 Linköping, Sweden; [email protected] (J.G.); [email protected] (M.R.-C.); [email protected] (C.A.M.); [email protected] (H.R.-M.) 2 Department of Animal Health and Anatomy, Veterinary Faculty, Universitat Autònoma de Barcelona, 08193 Bellaterra, Spain; [email protected] or [email protected] 3 Department of Physics, Chemistry and Biology, Faculty of Science and Engineering, Linköping University, 58185 Linköping, Sweden; [email protected] 4 College of Veterinary Medicine, Western University of Health Sciences, Pomona, CA 91766, USA * Correspondence: [email protected]; Tel.: +46-(0)729427883 Received: 10 June 2020; Accepted: 24 July 2020; Published: 27 July 2020 Abstract: The RNA-binding proteins (RBPs), some of them induced by transient receptor potential (TRP) ion channels, are crucial regulators of RNA function that can contribute to reproductive pathogenesis, including inflammation and immune dysfunction. This study aimed to reveal the influence of spermatozoa, seminal plasma, or natural mating on mRNA expression of RBPs and TRP ion channels in different segments of the internal genital tract of oestrous, preovulatory sows. -

Computational Analyses of Small Molecules Activity from Phenotypic Screens
Computational analyses of small molecules activity from phenotypic screens Azedine Zoufir Hughes Hall This dissertation is submitted for the degree of Doctor of Philosophy July 2018 Declaration This thesis is submitted as the result of my own work and includes nothing which is the outcome of work done in collaboration except where specifically indicated in the text. It is not substantially the same as any that I have submitted, or, is being concurrently submitted for a degree or diploma or other qualification at the University of Cambridge or any other University or similar institution except as declared in the preface and specified in the text. I further state that no substantial part of my dissertation has already been submitted, or, is being concurrently submitted for any such degree, diploma or other qualification at the University of Cambridge or any other University or similar institution except as declared in the Preface and specified in the text. This dissertation does not exceed the word limit of 60,000 words. Azedine Zoufir July 2018 Summary Title: Computational analyses of small molecules activity from phenotypic screens Author: Azedine Zoufir Drug discovery is no longer relying on the one gene-one disease paradigm nor on target-based screening alone to discover new drugs. Phenotypic-based screening is regaining momentum to discover new compounds since those assays provide an environment closer to the physiological state of the disease and allow to better anticipate off-target effects and other factors that can limit the efficacy of the drugs. However, uncovering the mechanism of action of the compounds active in those assays relies on in vitro techniques that are expensive and time- consuming. -

Genomic and Transcriptome Analysis Revealing an Oncogenic Functional Module in Meningiomas
Neurosurg Focus 35 (6):E3, 2013 ©AANS, 2013 Genomic and transcriptome analysis revealing an oncogenic functional module in meningiomas XIAO CHANG, PH.D.,1 LINGLING SHI, PH.D.,2 FAN GAO, PH.D.,1 JONATHAN RUssIN, M.D.,3 LIYUN ZENG, PH.D.,1 SHUHAN HE, B.S.,3 THOMAS C. CHEN, M.D.,3 STEVEN L. GIANNOTTA, M.D.,3 DANIEL J. WEISENBERGER, PH.D.,4 GAbrIEL ZADA, M.D.,3 KAI WANG, PH.D.,1,5,6 AND WIllIAM J. MAck, M.D.1,3 1Zilkha Neurogenetic Institute, Keck School of Medicine, University of Southern California, Los Angeles, California; 2GHM Institute of CNS Regeneration, Jinan University, Guangzhou, China; 3Department of Neurosurgery, Keck School of Medicine, University of Southern California, Los Angeles, California; 4USC Epigenome Center, Keck School of Medicine, University of Southern California, Los Angeles, California; 5Department of Psychiatry, Keck School of Medicine, University of Southern California, Los Angeles, California; and 6Division of Bioinformatics, Department of Preventive Medicine, Keck School of Medicine, University of Southern California, Los Angeles, California Object. Meningiomas are among the most common primary adult brain tumors. Although typically benign, roughly 2%–5% display malignant pathological features. The key molecular pathways involved in malignant trans- formation remain to be determined. Methods. Illumina expression microarrays were used to assess gene expression levels, and Illumina single- nucleotide polymorphism arrays were used to identify copy number variants in benign, atypical, and malignant me- ningiomas (19 tumors, including 4 malignant ones). The authors also reanalyzed 2 expression data sets generated on Affymetrix microarrays (n = 68, including 6 malignant ones; n = 56, including 3 malignant ones). -

Genomic Evidence of Reactive Oxygen Species Elevation in Papillary Thyroid Carcinoma with Hashimoto Thyroiditis
Endocrine Journal 2015, 62 (10), 857-877 Original Genomic evidence of reactive oxygen species elevation in papillary thyroid carcinoma with Hashimoto thyroiditis Jin Wook Yi1), 2), Ji Yeon Park1), Ji-Youn Sung1), 3), Sang Hyuk Kwak1), 4), Jihan Yu1), 5), Ji Hyun Chang1), 6), Jo-Heon Kim1), 7), Sang Yun Ha1), 8), Eun Kyung Paik1), 9), Woo Seung Lee1), Su-Jin Kim2), Kyu Eun Lee2)* and Ju Han Kim1)* 1) Division of Biomedical Informatics, Seoul National University College of Medicine, Seoul, Korea 2) Department of Surgery, Seoul National University Hospital and College of Medicine, Seoul, Korea 3) Department of Pathology, Kyung Hee University Hospital, Kyung Hee University School of Medicine, Seoul, Korea 4) Kwak Clinic, Okcheon-gun, Chungbuk, Korea 5) Department of Internal Medicine, Uijeongbu St. Mary’s Hospital, Uijeongbu, Korea 6) Department of Radiation Oncology, Seoul St. Mary’s Hospital, Seoul, Korea 7) Department of Pathology, Chonnam National University Hospital, Kwang-Ju, Korea 8) Department of Pathology, Samsung Medical Center, Sungkyunkwan University School of Medicine, Seoul, Korea 9) Department of Radiation Oncology, Korea Cancer Center Hospital, Korea Institute of Radiological and Medical Sciences, Seoul, Korea Abstract. Elevated levels of reactive oxygen species (ROS) have been proposed as a risk factor for the development of papillary thyroid carcinoma (PTC) in patients with Hashimoto thyroiditis (HT). However, it has yet to be proven that the total levels of ROS are sufficiently increased to contribute to carcinogenesis. We hypothesized that if the ROS levels were increased in HT, ROS-related genes would also be differently expressed in PTC with HT. To find differentially expressed genes (DEGs) we analyzed data from the Cancer Genomic Atlas, gene expression data from RNA sequencing: 33 from normal thyroid tissue, 232 from PTC without HT, and 60 from PTC with HT. -

Ohnologs in the Human Genome Are Dosage Balanced and Frequently Associated with Disease
Ohnologs in the human genome are dosage balanced and frequently associated with disease Takashi Makino1 and Aoife McLysaght2 Smurfit Institute of Genetics, University of Dublin, Trinity College, Dublin 2, Ireland Edited by Michael Freeling, University of California, Berkeley, CA, and approved April 9, 2010 (received for review December 21, 2009) About 30% of protein-coding genes in the human genome are been duplicated by WGD, subsequent loss of individual genes related through two whole genome duplication (WGD) events. would result in a dosage imbalance due to insufficient gene Although WGD is often credited with great evolutionary impor- product, thus leading to biased retention of dosage-balanced tance, the processes governing the retention of these genes and ohnologs. In fact, evidence for preferential retention of dosage- their biological significance remain unclear. One increasingly pop- balanced genes after WGD is accumulating (4, 7, 11–20). Copy ular hypothesis is that dosage balance constraints are a major number variation [copy number polymorphism (CNV)] describes determinant of duplicate gene retention. We test this hypothesis population level polymorphism of small segmental duplications and show that WGD-duplicated genes (ohnologs) have rarely and is known to directly correlate with gene expression levels (21– experienced subsequent small-scale duplication (SSD) and are also 24). Thus, CNV of dosage-balanced genes is also expected to be refractory to copy number variation (CNV) in human populations deleterious. This model predicts that retained ohnologs should be and are thus likely to be sensitive to relative quantities (i.e., they are enriched for dosage-balanced genes that are resistant to sub- dosage-balanced). -

Morphology, Behavior, and the Sonic Hedgehog Pathway in Mouse Models of Down Syndrome
MORPHOLOGY, BEHAVIOR, AND THE SONIC HEDGEHOG PATHWAY IN MOUSE MODELS OF DOWN SYNDROME by Tara Dutka A dissertation submitted to Johns Hopkins University in conformity with the requirements for the degree of Doctor of Philosophy Baltimore, Maryland July, 2014 © 2014 Tara Dutka All Rights Reserved Abstract Down Syndrome (DS) is caused by a triplication of human chromosome 21 (Hsa21). Ts65Dn, a mouse model of DS, contains a freely segregating extra chromosome consisting of the distal portion of mouse chromosome 16 (Mmu16), a region orthologous to part of Hsa21, and a non-Hsa21 orthologous region of mouse chromosome 17. All individuals with DS display some level of craniofacial dysmorphology, brain structural and functional changes, and cognitive impairment. Ts65Dn recapitulates these features of DS and aspects of each of these traits have been linked in Ts65Dn to a reduced response to Sonic Hedgehog (SHH) in trisomic cells. Dp(16)1Yey is a new mouse model of DS which has a direct duplication of the entire Hsa21 orthologous region of Mmu16. Dp(16)1Yey’s creators found similar behavioral deficits to those seen in Ts65Dn. We performed a quantitative investigation of the skull and brain of Dp(16)1Yey as compared to Ts65Dn and found that DS-like changes to brain and craniofacial morphology were similar in both models. Our results validate examination of the genetic basis for these phenotypes in Dp(16)1Yey mice and the genetic links for these phenotypes previously found in Ts65Dn , i.e., reduced response to SHH. Further, we hypothesized that if all trisomic cells show a reduced response to SHH, then up-regulation of the SHH pathway might ameliorate multiple phenotypes. -
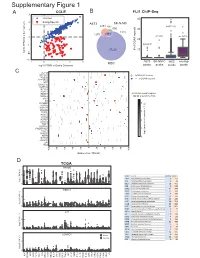
Supplementary Figure 1
6XSSOHPHQWDU\)LJXUH CCLE B FLI1 ChIP-Seq A 3 FRPPRQ 40 (ZLQJ6SHFLILF 2 A673 6.10& 477 Q 690 30 0 UHSHDWV Q Q A 20 í Q 57226 RI**$ ORJ)3.0LQRWKHUFDQFHUV í í A673 6.10& 06& RYHUODS í 0 23 06& ORJ)3.0LQ(ZLQJ6DUFRPD SHDNV SHDNV SHDNV SHDNV & ./) 67($3 (:6)/,ERXQG 3'(% ! **$AUHSHDWV 3335$ ABI3 '1$-& '863 13<5 .&1( 5%0 DOO**$AUHSHDWUHJLRQV 1.; NEDURXQGWKH766 9$9 ;* .&1$% 4 .&1$ 51) 0<20 3 355/ 355T4 &'$ $'5% 2 '&'& 711, 8*7$ 3+263+2 )(=) *1*7 ORJ QXPEHURIUHSHDWV A571 0 13<5 /2;+' /,3, 5$; 0 40 60 20 GLVWDQFHIURP766 NE D TCGA 35$0( 5 code cancer normal tumor LAML Acute Myeloid Leukemia 0 151 ORJ )3.0 ACC AdrenocoƌƟcal carcinoma 0 79 0 BLCA Bladder Urothelial Carcinoma 19 411 LGG Brain Lower Grade Glioma 0 529 COAD Colon adenocarcinoma 41 464 5%0 ESCA Esophageal carcinoma 11 162 GBM Glioblastoma muůƟforme 5 168 5 KICH Kidney Chromophobe 24 65 KIRP Kidney renal papillary cell carcinoma 32 289 LIHC Liver hepatocellular carcinoma 50 374 ORJ )3.0 LUAD Lung adenocarcinoma 59 526 0 LUSC Lung squamous cell carcinoma 49 501 DLBC Lymphoid Neoplasm Diīuse Large B-cell Lym 042 /,3, MESO Mesothelioma 0 1 OV Ovarian serous cystadenocarcinoma 0 379 PAAD PancreĂƟc adenocarcinoma 4 178 5 PCPG Pheochromocytoma and Paraganglioma 3 150 PRAD Prostate adenocarcinoma 52 499 READ Rectum adenocarcinoma 1 6 ORJ )3.0 SARC Sarcoma 0 35 0 SKCM Skin Cutaneous Melanoma 1 471 STAD Stomach adenocarcinoma 32 375 /2;+' WXPRU TGCT dĞƐƟĐƵůĂƌ'ĞƌŵĞůůdƵŵŽƌƐ 0156 THCA Thyroid carcinoma 58 510 QRUPDO UCS Uterine Carcinosarcoma 0 56 5 ORJ )3.0 0 / / 29 $&& 8&6 /** *%0