The Expression of Cold-Inducible RNA-Binding Protein Mrna in Sow Genital Tract Is Modulated by Natural Mating, but Not by Seminal Plasma
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Large-Scale Analysis of Genome and Transcriptome Alterations in Multiple Tumors Unveils Novel Cancer-Relevant Splicing Networks
Downloaded from genome.cshlp.org on September 28, 2021 - Published by Cold Spring Harbor Laboratory Press Research Large-scale analysis of genome and transcriptome alterations in multiple tumors unveils novel cancer-relevant splicing networks Endre Sebestyén,1,5 Babita Singh,1,5 Belén Miñana,1,2 Amadís Pagès,1 Francesca Mateo,3 Miguel Angel Pujana,3 Juan Valcárcel,1,2,4 and Eduardo Eyras1,4 1Universitat Pompeu Fabra, E08003 Barcelona, Spain; 2Centre for Genomic Regulation, E08003 Barcelona, Spain; 3Program Against Cancer Therapeutic Resistance (ProCURE), Catalan Institute of Oncology (ICO), Bellvitge Institute for Biomedical Research (IDIBELL), E08908 L’Hospitalet del Llobregat, Spain; 4Catalan Institution for Research and Advanced Studies, E08010 Barcelona, Spain Alternative splicing is regulated by multiple RNA-binding proteins and influences the expression of most eukaryotic genes. However, the role of this process in human disease, and particularly in cancer, is only starting to be unveiled. We system- atically analyzed mutation, copy number, and gene expression patterns of 1348 RNA-binding protein (RBP) genes in 11 solid tumor types, together with alternative splicing changes in these tumors and the enrichment of binding motifs in the alter- natively spliced sequences. Our comprehensive study reveals widespread alterations in the expression of RBP genes, as well as novel mutations and copy number variations in association with multiple alternative splicing changes in cancer drivers and oncogenic pathways. Remarkably, the altered splicing patterns in several tumor types recapitulate those of undifferen- tiated cells. These patterns are predicted to be mainly controlled by MBNL1 and involve multiple cancer drivers, including the mitotic gene NUMA1. We show that NUMA1 alternative splicing induces enhanced cell proliferation and centrosome am- plification in nontumorigenic mammary epithelial cells. -

RBM11 Sirna (H): Sc-91387
SANTA CRUZ BIOTECHNOLOGY, INC. RBM11 siRNA (h): sc-91387 BACKGROUND PRODUCT Proteins containing RNA recognition motifs, including various hnRNP proteins, RBM11 siRNA (h) is a pool of 3 target-specific 19-25 nt siRNAs designed are implicated in the regulation of alternative splicing and protein components to knock down gene expression. Each vial contains 3.3 nmol of lyophilized of snRNPs. The RBM (RNA-binding motif) gene family encodes proteins with siRNA, sufficient for a 10 µM solution once resuspended using protocol an RNA binding motif that have been suggested to play a role in the modu- below. Suitable for 50-100 transfections. Also see RBM11 shRNA Plasmid (h): lation of apoptosis. RBM11 (RNA-binding protein 11) is a 281 amino acid sc-91387-SH and RBM11 shRNA (h) Lentiviral Particles: sc-91387-V as alter- nuclear protein that contains one RNA recognition motif. RBM11 exists as nate gene silencing products. two isoforms produced by alternative splicing and is expressed in testis, kid- For independent verification of RBM11 (h) gene silencing results, we also ney, spleen, brain, spinal cord and mammary gland. The gene that encodes provide the individual siRNA duplex components. Each is available as 3.3 nmol RBM11 maps to chromosome 21, the smallest of the human chromosomes. of lyophilized siRNA. These include: sc-91387A, sc-91387B and sc-91387C. Down syndrome, also known as trisomy 21, is the disease most commonly associated with chromosome 21. Alzheimer’s disease, Jervell and Lange- STORAGE AND RESUSPENSION Nielsen syndrome and amyotrophic lateral sclerosis are also associated with chromosome 21. Store lyophilized siRNA duplex at -20° C with desiccant. -

Computational Analyses of Small Molecules Activity from Phenotypic Screens
Computational analyses of small molecules activity from phenotypic screens Azedine Zoufir Hughes Hall This dissertation is submitted for the degree of Doctor of Philosophy July 2018 Declaration This thesis is submitted as the result of my own work and includes nothing which is the outcome of work done in collaboration except where specifically indicated in the text. It is not substantially the same as any that I have submitted, or, is being concurrently submitted for a degree or diploma or other qualification at the University of Cambridge or any other University or similar institution except as declared in the preface and specified in the text. I further state that no substantial part of my dissertation has already been submitted, or, is being concurrently submitted for any such degree, diploma or other qualification at the University of Cambridge or any other University or similar institution except as declared in the Preface and specified in the text. This dissertation does not exceed the word limit of 60,000 words. Azedine Zoufir July 2018 Summary Title: Computational analyses of small molecules activity from phenotypic screens Author: Azedine Zoufir Drug discovery is no longer relying on the one gene-one disease paradigm nor on target-based screening alone to discover new drugs. Phenotypic-based screening is regaining momentum to discover new compounds since those assays provide an environment closer to the physiological state of the disease and allow to better anticipate off-target effects and other factors that can limit the efficacy of the drugs. However, uncovering the mechanism of action of the compounds active in those assays relies on in vitro techniques that are expensive and time- consuming. -

A Comparative Analysis of Transcribed Genes in the Mouse Hypothalamus and Neocortex Reveals Chromosomal Clustering
A comparative analysis of transcribed genes in the mouse hypothalamus and neocortex reveals chromosomal clustering Wee-Ming Boon*, Tim Beissbarth†, Lavinia Hyde†, Gordon Smyth†, Jenny Gunnersen*, Derek A. Denton*‡, Hamish Scott†, and Seong-Seng Tan* *Howard Florey Institute, University of Melbourne, Parkville 3052, Australia; and †Genetics and Bioinfomatics Division, Walter and Eliza Hall Institute of Medical Research, Royal Parade, Parkville 3050, Australia Contributed by Derek A. Denton, August 26, 2004 The hypothalamus and neocortex are subdivisions of the mamma- representing all of the genes that are expressed (qualitative and lian forebrain, and yet, they have vastly different evolutionary quantitative) in the hypothalamus and neocortex under standard histories, cytoarchitecture, and biological functions. In an attempt conditions. to define these attributes in terms of their genetic activity, we have In the current study, we describe the use of the Serial Analysis compared their genetic repertoires by using the Serial Analysis of of Gene Expression (SAGE) database, which allows simulta- Gene Expression database. From a comparison of 78,784 hypothal- neous detection of the expression levels of the entire genome amus tags with 125,296 neocortical tags, we demonstrate that each without a priori knowledge of gene sequences (13). SAGE takes structure possesses a different transcriptional profile in terms of advantage of the fact that a small sequence tag taken from a gene ontological characteristics and expression levels. Despite its defined position within the transcript is sufficient to identify the more recent evolutionary history, the neocortex has a more com- gene (from known cDNA or EST sequences), and up to 40 tags plex pattern of gene activity. -

Genomic Evidence of Reactive Oxygen Species Elevation in Papillary Thyroid Carcinoma with Hashimoto Thyroiditis
Endocrine Journal 2015, 62 (10), 857-877 Original Genomic evidence of reactive oxygen species elevation in papillary thyroid carcinoma with Hashimoto thyroiditis Jin Wook Yi1), 2), Ji Yeon Park1), Ji-Youn Sung1), 3), Sang Hyuk Kwak1), 4), Jihan Yu1), 5), Ji Hyun Chang1), 6), Jo-Heon Kim1), 7), Sang Yun Ha1), 8), Eun Kyung Paik1), 9), Woo Seung Lee1), Su-Jin Kim2), Kyu Eun Lee2)* and Ju Han Kim1)* 1) Division of Biomedical Informatics, Seoul National University College of Medicine, Seoul, Korea 2) Department of Surgery, Seoul National University Hospital and College of Medicine, Seoul, Korea 3) Department of Pathology, Kyung Hee University Hospital, Kyung Hee University School of Medicine, Seoul, Korea 4) Kwak Clinic, Okcheon-gun, Chungbuk, Korea 5) Department of Internal Medicine, Uijeongbu St. Mary’s Hospital, Uijeongbu, Korea 6) Department of Radiation Oncology, Seoul St. Mary’s Hospital, Seoul, Korea 7) Department of Pathology, Chonnam National University Hospital, Kwang-Ju, Korea 8) Department of Pathology, Samsung Medical Center, Sungkyunkwan University School of Medicine, Seoul, Korea 9) Department of Radiation Oncology, Korea Cancer Center Hospital, Korea Institute of Radiological and Medical Sciences, Seoul, Korea Abstract. Elevated levels of reactive oxygen species (ROS) have been proposed as a risk factor for the development of papillary thyroid carcinoma (PTC) in patients with Hashimoto thyroiditis (HT). However, it has yet to be proven that the total levels of ROS are sufficiently increased to contribute to carcinogenesis. We hypothesized that if the ROS levels were increased in HT, ROS-related genes would also be differently expressed in PTC with HT. To find differentially expressed genes (DEGs) we analyzed data from the Cancer Genomic Atlas, gene expression data from RNA sequencing: 33 from normal thyroid tissue, 232 from PTC without HT, and 60 from PTC with HT. -

An Annual Cycle of Gene Regulation in the Red-Legged Salamander Mental Gland: from Hypertrophy to Expression of Rapidly Evolving Pheromones
bioRxiv preprint doi: https://doi.org/10.1101/261495; this version posted February 7, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. An annual cycle of gene regulation in the red-legged salamander mental gland: from hypertrophy to expression of rapidly evolving pheromones Damien B. Wilburn1,2§ and Richard C. Feldhoff1 1Department of Biochemistry and Molecular Biology, University of Louisville, Louisville, KY 40292 2Department of Genome Sciences, University of Washington, Seattle, WA 98195 §Corresponding author Email addresses: DBW: [email protected] RCF: [email protected] - 1 - bioRxiv preprint doi: https://doi.org/10.1101/261495; this version posted February 7, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. Abstract Cell differentiation is mediated by synchronized waves of coordinated expression for hundreds to thousands of genes, and must be an exquisitely regulated process to produce complex tissues and phenotypes. For many animal species, sexual selection has driven the development of elaborate male ornaments, requiring sex-specific differentiation pathways. One such male ornament is the pheromone-producing mental gland of the red-legged salamander (Plethodon shermani). Mental gland development follows an annual cycle of extreme hypertrophy, production of pheromones for the ~2 month mating season, and then complete resorption before repeating the process in the following year. -

Ohnologs in the Human Genome Are Dosage Balanced and Frequently Associated with Disease
Ohnologs in the human genome are dosage balanced and frequently associated with disease Takashi Makino1 and Aoife McLysaght2 Smurfit Institute of Genetics, University of Dublin, Trinity College, Dublin 2, Ireland Edited by Michael Freeling, University of California, Berkeley, CA, and approved April 9, 2010 (received for review December 21, 2009) About 30% of protein-coding genes in the human genome are been duplicated by WGD, subsequent loss of individual genes related through two whole genome duplication (WGD) events. would result in a dosage imbalance due to insufficient gene Although WGD is often credited with great evolutionary impor- product, thus leading to biased retention of dosage-balanced tance, the processes governing the retention of these genes and ohnologs. In fact, evidence for preferential retention of dosage- their biological significance remain unclear. One increasingly pop- balanced genes after WGD is accumulating (4, 7, 11–20). Copy ular hypothesis is that dosage balance constraints are a major number variation [copy number polymorphism (CNV)] describes determinant of duplicate gene retention. We test this hypothesis population level polymorphism of small segmental duplications and show that WGD-duplicated genes (ohnologs) have rarely and is known to directly correlate with gene expression levels (21– experienced subsequent small-scale duplication (SSD) and are also 24). Thus, CNV of dosage-balanced genes is also expected to be refractory to copy number variation (CNV) in human populations deleterious. This model predicts that retained ohnologs should be and are thus likely to be sensitive to relative quantities (i.e., they are enriched for dosage-balanced genes that are resistant to sub- dosage-balanced). -

Human CIRBP (1-172, His-Tag) - Purified
OriGene Technologies, Inc. OriGene Technologies GmbH 9620 Medical Center Drive, Ste 200 Schillerstr. 5 Rockville, MD 20850 32052 Herford UNITED STATES GERMANY Phone: +1-888-267-4436 Phone: +49-5221-34606-0 Fax: +1-301-340-8606 Fax: +49-5221-34606-11 [email protected] [email protected] AR09973PU-N Human CIRBP (1-172, His-tag) - Purified Alternate names: A18HNRNP, CIRP, Cold-inducible RNA-binding protein, Glycine-rich RNA-binding protein CIRP Quantity: 0.1 mg Concentration: 0.5 mg/ml (determined by Bradford assay) Background: Cold inducible RNA-binding protein, also known as CIRBP, plays a protective role in the genotoxic stress response by stabilizing transcripts of genes involved in cell survival. It is localizes to the nucleus and contains one RRM (RNA recognition motif) domain. Expressed ubiquitously, CIRBP is thought to play an essential role in the suppression of cellular proliferation in response to UV irradiation or extreme cold. Uniprot ID: Q14011 NCBI: NP_001271 GeneID: 1153 Species: Human Source: E. coli Format: State: Liquid purified protein Purity: >95% by SDS-PAGE Buffer System: 20mM Tris-HCl buffer (pH 8.0) containing 1mM DTT, 50% glycerol, 0.2M NaCl Description: Recombinant human CIRBP protein, fused to His-tag at N-terminus, was expressed in E.coli and purified by using conventional chromatography techniques. AA Sequence: MGSSHHHHHH SSGLVPRGSH MASDEGKLFV GGLSFDTNEQ SLEQVFSKYG QISEVVVVKD RETQRSRGFG FVTFENIDDA KDAMMAMNGK SVDGRQIRVD QAGKSSDNRS RGYRGGSAGG RGFFRGGRGR GRGFSRGGGD RGYGGNRFES RSGGYGGSRD YYSSRSQSGG YSDRSSGGSY RDSYDSYATH NE Molecular weight: 20.8 kDa (192aa) confirmed by MALDI-TOF Storage: Store undiluted at 2-8°C for up to two weeks or (in aliquots) at -20°C or -70°C for longer. -

CIRBP Polyclonal Antibody
PRODUCT DATA SHEET Bioworld Technology,Inc. CIRBP polyclonal antibody Catalog: BS6943 Host: Rabbit Reactivity: Human,Mouse,Rat BackGround: by affinity-chromatography using epitope-specific im- CIRP, also known as CIRBP (cold inducible RNA binding munogen and the purity is > 95% (by SDS-PAGE). protein) or A18HNRNP, is a 172 amino acid protein that Applications: localizes to the nucleus and contains one RRM (RNA WB: 1:500~1:2000 recognition motif) domain. Expressed ubiquitously, CIRP IHC: 1:50~1:200 is thought to play an essential role in the suppression of Storage&Stability: cellular proliferation in response to UV irradiation or ex- Store at 4°C short term. Aliquot and store at -20°C long treme cold. Human CIRP, which may be involved in the term. Avoid freeze-thaw cycles. pathogenesis of endometrial carcinoma, shares 95% se- Specificity: quence identity with its mouse counterpart, suggesting a CIRBP polyclonal antibody detects endogenous levels of conserved role between species. The gene encoding CIRP CIRBP protein. maps to human chromosome 19, which is the genetic DATA: home for a number of immunoglobulin superfamily members, including the killer cell and leukocyte Ig-like receptors, a number of ICAMs, the CEACAM and PSG family and Fc receptors (FcRs). Product: Rabbit IgG, 1mg/ml in PBS with 0.02% sodium azide, 50% glycerol, pH7.2 Molecular Weight: WesternBlot (WB) analysis of CIRBP polyclonal antibody ~ 19 kDa Note: Swiss-Prot: For research use only, not for use in diagnostic procedure. Q14011 Purification&Purity: The antibody was affinity-purified from rabbit antiserum Bioworld Technology, Inc. Bioworld technology, co. Ltd. -

Morphology, Behavior, and the Sonic Hedgehog Pathway in Mouse Models of Down Syndrome
MORPHOLOGY, BEHAVIOR, AND THE SONIC HEDGEHOG PATHWAY IN MOUSE MODELS OF DOWN SYNDROME by Tara Dutka A dissertation submitted to Johns Hopkins University in conformity with the requirements for the degree of Doctor of Philosophy Baltimore, Maryland July, 2014 © 2014 Tara Dutka All Rights Reserved Abstract Down Syndrome (DS) is caused by a triplication of human chromosome 21 (Hsa21). Ts65Dn, a mouse model of DS, contains a freely segregating extra chromosome consisting of the distal portion of mouse chromosome 16 (Mmu16), a region orthologous to part of Hsa21, and a non-Hsa21 orthologous region of mouse chromosome 17. All individuals with DS display some level of craniofacial dysmorphology, brain structural and functional changes, and cognitive impairment. Ts65Dn recapitulates these features of DS and aspects of each of these traits have been linked in Ts65Dn to a reduced response to Sonic Hedgehog (SHH) in trisomic cells. Dp(16)1Yey is a new mouse model of DS which has a direct duplication of the entire Hsa21 orthologous region of Mmu16. Dp(16)1Yey’s creators found similar behavioral deficits to those seen in Ts65Dn. We performed a quantitative investigation of the skull and brain of Dp(16)1Yey as compared to Ts65Dn and found that DS-like changes to brain and craniofacial morphology were similar in both models. Our results validate examination of the genetic basis for these phenotypes in Dp(16)1Yey mice and the genetic links for these phenotypes previously found in Ts65Dn , i.e., reduced response to SHH. Further, we hypothesized that if all trisomic cells show a reduced response to SHH, then up-regulation of the SHH pathway might ameliorate multiple phenotypes. -
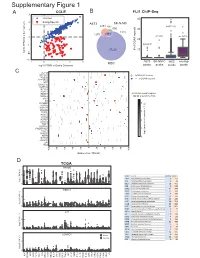
Supplementary Figure 1
6XSSOHPHQWDU\)LJXUH CCLE B FLI1 ChIP-Seq A 3 FRPPRQ 40 (ZLQJ6SHFLILF 2 A673 6.10& 477 Q 690 30 0 UHSHDWV Q Q A 20 í Q 57226 RI**$ ORJ)3.0LQRWKHUFDQFHUV í í A673 6.10& 06& RYHUODS í 0 23 06& ORJ)3.0LQ(ZLQJ6DUFRPD SHDNV SHDNV SHDNV SHDNV & ./) 67($3 (:6)/,ERXQG 3'(% ! **$AUHSHDWV 3335$ ABI3 '1$-& '863 13<5 .&1( 5%0 DOO**$AUHSHDWUHJLRQV 1.; NEDURXQGWKH766 9$9 ;* .&1$% 4 .&1$ 51) 0<20 3 355/ 355T4 &'$ $'5% 2 '&'& 711, 8*7$ 3+263+2 )(=) *1*7 ORJ QXPEHURIUHSHDWV A571 0 13<5 /2;+' /,3, 5$; 0 40 60 20 GLVWDQFHIURP766 NE D TCGA 35$0( 5 code cancer normal tumor LAML Acute Myeloid Leukemia 0 151 ORJ )3.0 ACC AdrenocoƌƟcal carcinoma 0 79 0 BLCA Bladder Urothelial Carcinoma 19 411 LGG Brain Lower Grade Glioma 0 529 COAD Colon adenocarcinoma 41 464 5%0 ESCA Esophageal carcinoma 11 162 GBM Glioblastoma muůƟforme 5 168 5 KICH Kidney Chromophobe 24 65 KIRP Kidney renal papillary cell carcinoma 32 289 LIHC Liver hepatocellular carcinoma 50 374 ORJ )3.0 LUAD Lung adenocarcinoma 59 526 0 LUSC Lung squamous cell carcinoma 49 501 DLBC Lymphoid Neoplasm Diīuse Large B-cell Lym 042 /,3, MESO Mesothelioma 0 1 OV Ovarian serous cystadenocarcinoma 0 379 PAAD PancreĂƟc adenocarcinoma 4 178 5 PCPG Pheochromocytoma and Paraganglioma 3 150 PRAD Prostate adenocarcinoma 52 499 READ Rectum adenocarcinoma 1 6 ORJ )3.0 SARC Sarcoma 0 35 0 SKCM Skin Cutaneous Melanoma 1 471 STAD Stomach adenocarcinoma 32 375 /2;+' WXPRU TGCT dĞƐƟĐƵůĂƌ'ĞƌŵĞůůdƵŵŽƌƐ 0156 THCA Thyroid carcinoma 58 510 QRUPDO UCS Uterine Carcinosarcoma 0 56 5 ORJ )3.0 0 / / 29 $&& 8&6 /** *%0 -

Variation in Protein Coding Genes Identifies Information Flow
bioRxiv preprint doi: https://doi.org/10.1101/679456; this version posted June 21, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. Animal complexity and information flow 1 1 2 3 4 5 Variation in protein coding genes identifies information flow as a contributor to 6 animal complexity 7 8 Jack Dean, Daniela Lopes Cardoso and Colin Sharpe* 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 Institute of Biological and Biomedical Sciences 25 School of Biological Science 26 University of Portsmouth, 27 Portsmouth, UK 28 PO16 7YH 29 30 * Author for correspondence 31 [email protected] 32 33 Orcid numbers: 34 DLC: 0000-0003-2683-1745 35 CS: 0000-0002-5022-0840 36 37 38 39 40 41 42 43 44 45 46 47 48 49 Abstract bioRxiv preprint doi: https://doi.org/10.1101/679456; this version posted June 21, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. Animal complexity and information flow 2 1 Across the metazoans there is a trend towards greater organismal complexity. How 2 complexity is generated, however, is uncertain. Since C.elegans and humans have 3 approximately the same number of genes, the explanation will depend on how genes are 4 used, rather than their absolute number.