Antigen Processing and Presentation in Cancer Immunotherapy
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Antigen Presentation by Dendritic Cells and Their Significance in Antineoplastic Immunotherapy
in vivo 18: 81-100 (2004) Antigen Presentation by Dendritic Cells and their Significance in Antineoplastic Immunotherapy BELA BODEY1,2, STUART E. SIEGEL1,2 and HANS E. KAISER3 1Department of Pathology, Keck School of Medicine, University of Southern California, Los Angeles, CA; 2Childrens’ Center for Cancer and Blood Diseases, Childrens’ Hospital Los Angeles, Los Angeles, CA; 3Department of Pathology, School of Medicine, University of Maryland, Baltimore, MD, U.S.A. and Department of General and Experimental Pathology, University of Vienna, Vienna, Austria Abstract. Dendritic cells (DCs) are present in essentially every derived peptides on the surface of major histocompatibility mammalian tissue, where they operate at the interface of innate complex (MHC) molecules and acquire the cellular specialization and acquired immunity by recognizing pathogens and presenting to select and activate naive antigen-specific T lymphocytes. pathogen-derived peptides to T lymphocytes. According to the Immunotherapeutic ideas are based on the ability of the research group of Shortman (1-9), experimental results suggest a mammalian immune system to recognize neoplastically "dual" DC differentiation model, demonstrating the existence of transformed cells. Immunotherapy of human neoplasms has both myeloid-derived (with characteristic IF: CD11b+, CD11c+, always represented a very attractive fourth-modality therapeutic CD8alpha- and DEC205+) and lymphoid-derived DCs (showing approach, especially in light of the many shortcomings of CD11b-, CD11c-, CD8alpha+ and DEC205+ IF). DCs, including conventional surgical, radiation and chemotherapies in the interdigitating cells (IDCs) and Langerhans cells (LCs), are management of neoplastically transformed cells. The cancer characterized by dendritic morphology, high migratory mobility and vaccine approach to therapy is based on the notion that the are the most effective, "professional" cells for antigen presentation immune system could possibly mount a rejection strength response in primary immune responses. -

"Epitope Mapping: B-Cell Epitopes". In: Encyclopedia of Life Sciences
Epitope Mapping: B-cell Advanced article Epitopes Article Contents . Introduction GE Morris, Wolfson Centre for Inherited Neuromuscular Disease RJAH Orthopaedic Hospital, . What Is a B-cell Epitope? . Epitope Mapping Methods Oswestry, UK and Keele University, Keele, Staffordshire, UK . Applications Immunoglobulin molecules are folded to present a surface structure complementary to doi: 10.1002/9780470015902.a0002624.pub2 a surface feature on the antigen – the epitope is this feature of the antigen. Epitope mapping is the process of locating the antibody-binding site on the antigen, although the term is also applied more broadly to receptor–ligand interactions unrelated to the immune system. Introduction formed of highly convoluted peptide chains, so that resi- dues that lie close together on the protein surface are often Immunoglobulin molecules are folded in a way that as- far apart in the amino acid sequence (Barlow et al., 1986). sembles sequences from the variable regions of both the Consequently, most epitopes on native, globular proteins heavy and light chains into a surface feature (comprised of are conformation-dependent and they disappear if the up to six complementarity-determining regions (CDRs)) protein is denatured or fragmented. Sometimes, by acci- that is complementary in shape to a surface structure on the dent or design, antibodies are produced against linear antigen. These two surface features, the ‘paratope’ on the (sequential) epitopes that survive denaturation, though antibody and the ‘epitope’ on the antigen, may have a cer- such antibodies usually fail to recognize the native protein. tain amount of flexibility to allow an ‘induced fit’ between The simplest way to find out whether an epitope is confor- them. -

Antigen Processing (Major Hbitocompatlbility Complex/Class I Moecules/Lymphokines) YOUNG YANG, JAMES B
Proc. Nati. Acad. Sci. USA Vol. 89, pp. 4928-4932, June 1992 Immunology Proteasomes are regulated by interferon y: Implications for antigen processing (major hbitocompatlbility complex/class I moecules/lymphokines) YOUNG YANG, JAMES B. WATERS, KLAUS FROH, AND PER A. PETERSON Department of Immunology, The Scripps Research Institute, La Jolla, CA 92037 Communicated by Frank J. Dixon, February 27, 1992 ABSTRACT Class I major histocompatibility complex strengthened by the findings, presented in this communica- (MHC) molecules present antigenic peptides of cytoplasmic tion, that several proteasomal subunits, including MHC- origin to T cells. As the lengths ofthese peptides seem stried encoded subunits, are regulated by interferon y (IFN--y) and to eight or nine amino acids, an unusual proteolytic system that the incorporation of several more subunits into protea- must play a role in antigen processing. Proteasomes, a major somes appears to depend on the expression of the MHC- extralysosomal proteolytic system, are responsible for the encoded proteasomal subunits. Moreover, the pattern of degradation of cytoplasmic proteins. We demonstrate that expression of IFN-y-regulated subunits suggests complexi- several proteasomal subunits, including MHC-encoded sub- ties in the regulation of proteasomes with respect to its units, are regulated by interferon y. These data and the finding subunit composition, subcellular localization, and its incor- that MHC-encoded and other interferon -regulated protea- poration into larger ubiquitin-related proteolytic complexes. somal subunits are uniquely associated with proteasomes Possible functions for the MHC-encoded and IFN-y- strongly suggest that the immune system has recruited protea- regulated proteasomal subunits in antigen processing are somes for antigen processing. -

Cytoplasmic Proteolysis Antigens on MHC Class II Is Dependent On
Efficient Presentation of Both Cytosolic and Endogenous Transmembrane Protein Antigens on MHC Class II Is Dependent on Cytoplasmic Proteolysis This information is current as of September 28, 2021. Paushali Mukherjee, Aadish Dani, Sumeena Bhatia, Nagendra Singh, Alexander Y. Rudensky, Anna George, Vineeta Bal, Satyajit Mayor and Satyajit Rath J Immunol 2001; 167:2632-2641; ; doi: 10.4049/jimmunol.167.5.2632 Downloaded from http://www.jimmunol.org/content/167/5/2632 References This article cites 51 articles, 20 of which you can access for free at: http://www.jimmunol.org/content/167/5/2632.full#ref-list-1 http://www.jimmunol.org/ Why The JI? Submit online. • Rapid Reviews! 30 days* from submission to initial decision • No Triage! Every submission reviewed by practicing scientists by guest on September 28, 2021 • Fast Publication! 4 weeks from acceptance to publication *average Subscription Information about subscribing to The Journal of Immunology is online at: http://jimmunol.org/subscription Permissions Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html Email Alerts Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts The Journal of Immunology is published twice each month by The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2001 by The American Association of Immunologists All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. Efficient Presentation of Both Cytosolic and Endogenous Transmembrane Protein Antigens on MHC Class II Is Dependent on Cytoplasmic Proteolysis1 Paushali Mukherjee,* Aadish Dani,† Sumeena Bhatia,* Nagendra Singh,* Alexander Y. -

Epstein-Barr Virus Epitope-Major Histocompatibility Complex
University of Massachusetts Medical School eScholarship@UMMS Open Access Articles Open Access Publications by UMMS Authors 2020-03-17 Epstein-Barr Virus Epitope-Major Histocompatibility Complex Interaction Combined with Convergent Recombination Drives Selection of Diverse T Cell Receptor alpha and beta Repertoires Anna Gil University of Massachusetts Medical School Et al. Let us know how access to this document benefits ou.y Follow this and additional works at: https://escholarship.umassmed.edu/oapubs Part of the Hemic and Lymphatic Diseases Commons, Immune System Diseases Commons, Immunology and Infectious Disease Commons, Infectious Disease Commons, Microbiology Commons, Virus Diseases Commons, and the Viruses Commons Repository Citation Gil A, Kamga L, Chirravuri-Venkata R, Aslan N, Clark FG, Ghersi D, Luzuriaga K, Selin LK. (2020). Epstein- Barr Virus Epitope-Major Histocompatibility Complex Interaction Combined with Convergent Recombination Drives Selection of Diverse T Cell Receptor alpha and beta Repertoires. Open Access Articles. https://doi.org/10.1128/mBio.00250-20. Retrieved from https://escholarship.umassmed.edu/ oapubs/4191 Creative Commons License This work is licensed under a Creative Commons Attribution 4.0 License. This material is brought to you by eScholarship@UMMS. It has been accepted for inclusion in Open Access Articles by an authorized administrator of eScholarship@UMMS. For more information, please contact [email protected]. RESEARCH ARTICLE Host-Microbe Biology crossm Epstein-Barr Virus Epitope–Major Histocompatibility Complex Interaction Combined with Convergent Recombination Drives Downloaded from Selection of Diverse T Cell Receptor ␣ and  Repertoires Anna Gil,a Larisa Kamga,b Ramakanth Chirravuri-Venkata,c Nuray Aslan,a Fransenio Clark,a Dario Ghersi,c Katherine Luzuriaga,b Liisa K. -

Epitope Spreading: Lessons from Autoimmune Skin Diseases
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by Elsevier - Publisher Connector REVIEW Epitope Spreading: Lessons From Autoimmune Skin Diseases Lawrence S. Chan,*† Carol J. Vanderlugt,‡ Takashi Hashimoto,§ Takeji Nishikawa,¶ John J. Zone,** Martin M. Black,†† Fenella Wojnarowska,‡‡ Seth R. Stevens,§§ Mei Chen,† Janet A. Fairley,¶¶ David T. Woodley,*† Stephen D. Miller,‡ and Kenneth B. Gordon†‡ *Medicine Service, Section of Dermatology, Lakeside Division, VA Chicago Health Care System, Chicago, Illinois, U.S.A.; Departments of †Dermatology and ‡Microbiology and Immunology, Northwestern University Medical School, Chicago, Illinois, U.S.A.; ¶¶Department of Dermatology, Kurume University School of Medicine, Kurume, Japan; ¶Department of Dermatology, Keio University School of Medicine, Tokyo, Japan; **Medicine Service, Section of Dermatology, Salt Lake City VA Medical Center, Salt Lake City, Utah, U.S.A.; ††Department of Dermatopathology, Guy’s and St. Thomas Medical and Dental School, London, U.K.; ‡‡Department of Dermatology, The Oxford Radcliffe Hospital, Oxford, U.K.; §§Department of Dermatology, Case Western Reserve University School of Medicine, Cleveland, Ohio, U.S.A.; ¶¶Department of Dermatology, Medical College of Wisconsin, Milwaukee, Wisconsin, U.S.A. Autoimmune diseases are initiated when patients develop In experimental autoimmune animal diseases, ‘‘epitope aberrant T and/or B cell responses against self proteins. spreading’’ seems to have significant physiologic impor- These responses -
Antigen Processing and Presentation by Mhcs Pathway
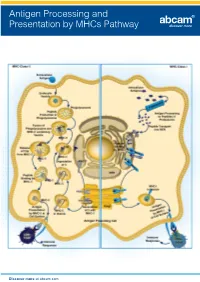
Antigen Processing and Presentation by MHCs Pathway Discover more at abcam.com Antigen Processing and Presentation by MHCs Pathway Related antibodies Product This image shows MHC Class II highlights Product Clonality Applications Host Species Reactivity Product code expressing cells in formalin fixed and paraffin embedded human lymph MHC class I antibody [EP1395Y] M Flow Cyt, ICC/IF, IHC-P, IP, WB Rb Hu, Ms, Rat 52922 node, using ab55152. MHC class I antibody [AF6-88.5.5.3] (Biotin) M Flow Cyt Ms Ms 93528 MHC Class II antibody MHC class I antibody [34-1-2S] (FITC) M Flow Cyt Ms Ms 95572 (ab55152) MHC class I antibody [34-1-2S] (Phycoerythrin) M Flow Cyt Ms Ms 95571 MHC Class I alpha antibody [F21-2] M AP, Flow Cyt, IP, WB Ms Chk 23483 Clonality Applications MHC Class 1 H2 Db antibody [27-11-13] - BSA and Azide free M Flow Cyt, FuncS, IHC-Fr Ms Ms 25244 M IHC-P, WB MHC Class 1 H2 Db antibody [28-14-8] (FITC) M Flow Cyt Ms Ms 25056 Host Species cross reactivity MHC Class 1 H2 Db antibody [KH95] M Flow Cyt, FuncS, WB Ms Ms 64373 Ms Hu, RecFrag MHC Class I H2 Dd antibody [34-2-12] M Flow Cyt, FuncS, IHC-Fr, IP Ms Ms 64368 MHC Class I H2 Dk antibody [15-5-5.3] M Flow Cyt Ms Ms 25216 MHC Class II antigens are a valuable tool for studying T helper cell interactions with class II MHC Class I H2 Kb antibody P ELISA, WB Rb Ms 93364 positive antigen presenting cells, as well as the MHC Class I H2 Kd + Dd + q + u + v antibody [1.B.552] (FITC) M Flow Cyt, IP Ms Ms 62228 development of T helper cells since the antigen is present on stromal cells in the thymus. -

A Murine CD8+ T Cell Epitope Identified in the Receptor-Binding
Article A Murine CD8+ T Cell Epitope Identified in the Receptor-Binding Domain of the SARS-CoV-2 Spike Protein Jihyun Yang 1,† , Eunjin Kim 1,2,†, Jong-Soo Lee 2 and Haryoung Poo 1,* 1 Infectious Disease Research Center, Korea Research Institute of Bioscience and Biotechnology (KRIBB), Daejeon 34141, Korea; [email protected] (J.Y.); [email protected] (E.K.) 2 Department of Preventive Veterinary Medicine, College of Veterinary Medicine, Chungnam National University, Daejeon 34134, Korea; [email protected] * Correspondence: [email protected]; Tel.: +82-42-860-4157 † These authors contributed equally to this study. Abstract: The ongoing COVID-19 pandemic caused by SARS-CoV-2 has posed a devastating threat worldwide. The receptor-binding domain (RBD) of the spike protein is one of the most important antigens for SARS-CoV-2 vaccines, while the analysis of CD8 cytotoxic T lymphocyte activity in preclinical studies using mouse models is critical for evaluating vaccine efficacy. Here, we immunized C57BL/6 wild-type mice and transgenic mice expressing human angiotensin-converting enzyme 2 (ACE2) with the SARS-CoV-2 RBD protein to evaluate the IFN-γ-producing T cells in the splenocytes of the immunized mice using an overlapping peptide pool by an enzyme-linked immunospot assay and flow cytometry. We identified SARS-CoV-2 S395–404 as a major histocompatibility complex (MHC) class I-restricted epitope for the RBD-specific CD8 T cell responses in C57BL/6 mice. Keywords: SARS-CoV-2; cell-mediated immunity; CD8 cytotoxic T lymphocyte; epitope; vaccine Citation: Yang, J.; Kim, E.; Lee, J.-S.; Poo, H. -

Heteroclitic CD8 T Cell Epitopes Enhanced Antiviral Immunity
Structural and Functional Correlates of Enhanced Antiviral Immunity Generated by Heteroclitic CD8 T Cell Epitopes This information is current as Jonathan A. Trujillo, Stephanie Gras, Kelly-Anne Twist, of September 30, 2021. Nathan P. Croft, Rudragouda Channappanavar, Jamie Rossjohn, Anthony W. Purcell and Stanley Perlman J Immunol 2014; 192:5245-5256; Prepublished online 2 May 2014; doi: 10.4049/jimmunol.1400111 Downloaded from http://www.jimmunol.org/content/192/11/5245 References This article cites 56 articles, 31 of which you can access for free at: http://www.jimmunol.org/content/192/11/5245.full#ref-list-1 http://www.jimmunol.org/ Why The JI? Submit online. • Rapid Reviews! 30 days* from submission to initial decision • No Triage! Every submission reviewed by practicing scientists by guest on September 30, 2021 • Fast Publication! 4 weeks from acceptance to publication *average Subscription Information about subscribing to The Journal of Immunology is online at: http://jimmunol.org/subscription Permissions Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html Email Alerts Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts The Journal of Immunology is published twice each month by The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2014 by The American Association of Immunologists, Inc. All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. The Journal of Immunology Structural and Functional Correlates of Enhanced Antiviral Immunity Generated by Heteroclitic CD8 T Cell Epitopes Jonathan A. Trujillo,*,1 Stephanie Gras,†,1 Kelly-Anne Twist,† Nathan P. -

Mechanisms of HIV Protein Degradation Into Epitopes: Implications for Vaccine Design
Viruses 2014, 6, 3271-3292; doi:10.3390/v6083271 OPEN ACCESS viruses ISSN 1999-4915 www.mdpi.com/journal/viruses Review Mechanisms of HIV Protein Degradation into Epitopes: Implications for Vaccine Design Marijana Rucevic †, Julie Boucau †, Jens Dinter †, Georgio Kourjian † and Sylvie Le Gall * Ragon Institute of MGH, MIT and Harvard, Massachusetts General Hospital and Harvard Medical School, Cambridge, MA 02139, USA; E-Mails: [email protected] (M.R.); [email protected] (J.B.); [email protected] (J.D.); [email protected] (G.K.) † These authors contributed equally to this work. * Author to whom correspondence should be addressed; E-Mail: [email protected]; Tel.: +1-857-268-7010. Received: 11 June 2014; in revised form: 6 August 2014 / Accepted: 11 August 2014 / Published: 21 August 2014 Abstract: The degradation of HIV-derived proteins into epitopes displayed by MHC-I or MHC-II are the first events leading to the priming of HIV-specific immune responses and to the recognition of infected cells. Despite a wealth of information about peptidases involved in protein degradation, our knowledge of epitope presentation during HIV infection remains limited. Here we review current data on HIV protein degradation linking epitope production and immunodominance, viral evolution and impaired epitope presentation. We propose that an in-depth understanding of HIV antigen processing and presentation in relevant primary cells could be exploited to identify signatures leading to efficient or inefficient epitope presentation in HIV proteomes, and to improve the design of immunogens eliciting immune responses efficiently recognizing all infected cells. Keywords: HIV; antigen processing; protein degradation; proteasome; aminopeptidase; peptidase; immunogen; vaccine vector; dendritic cells; T cells; viral evolution Viruses 2014, 6 3272 1. -

Epitope Similarity Cannot Explain the Pre-Formed T Cell Immunity
www.nature.com/scientificreports OPEN Epitope similarity cannot explain the pre‑formed T cell immunity towards structural SARS‑CoV‑2 proteins Ulrik Stervbo 1,2,4*, Sven Rahmann 3,4*, Toralf Roch 1,2, Timm H. Westhof1 & Nina Babel1,2 The current pandemic is caused by the SARS‑CoV‑2 virus and large progress in understanding the pathology of the virus has been made since its emergence in late 2019. Several reports indicate short lasting immunity against endemic coronaviruses, which contrasts studies showing that biobanked venous blood contains T cells reactive to SARS‑CoV‑2 S‑protein even before the outbreak in Wuhan. This suggests a preformed T cell memory towards structural proteins in individuals not exposed to SARS‑CoV‑2. Given the similarity of SARS‑CoV‑2 to other members of the Coronaviridae family, the endemic coronaviruses appear likely candidates to generate this T cell memory. However, given the apparent poor immunological memory created by the endemic coronaviruses, immunity against other common pathogens might ofer an alternative explanation. Here, we utilize a combination of epitope prediction and similarity to common human pathogens to identify potential sources of the SARS‑CoV‑2 T cell memory. Although beta‑coronaviruses are the most likely candidates to explain the pre‑existing SARS‑CoV‑2 reactive T cells in uninfected individuals, the SARS‑CoV‑2 epitopes with the highest similarity to those from beta‑coronaviruses are confned to replication associated proteins—not the host interacting S‑protein. Thus, our study suggests that the observed SARS‑CoV‑2 pre‑formed immunity to structural proteins is not driven by near‑identical epitopes. -

Exploring Epitope and Functional Diversity of Anti-SARS-Cov2 Antibodies Using AI-Based Methods
bioRxiv preprint doi: https://doi.org/10.1101/2020.12.23.424199; this version posted December 24, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Exploring epitope and functional diversity of anti-SARS-CoV2 antibodies using AI-based methods. Dumet C1., Jullian Y.1, Musnier A.1, Rivière Ph.2, Poirier N.3, Watier H.4, Bourquard T.1, Poupon A.1,5,* 1: MAbSilico SAS, 1 impasse du Palais, 37000 Tours, France. 2: VisionsCarto, France. 3: OSE Immunotherapeutics, Nantes, France. 4: EA7501, Université de Tours, et laboratoire d'immunologie, CHU de Tours. 5: PRC, INRAE, CNRS, Université de Tours, 37380 Nouzilly, France ; Inria, Inria Saclay-Île-de-France, 91120 Palaiseau, France *: corresponding author. Summary Since the beginning of the COVID19 pandemics, an unprecedented research effort has been conducted to analyze the antibody responses in patients, and many trials based on passive immunotherapy — notably monoclonal antibodies — are ongoing. Twenty-one antibodies have entered clinical trials, 6 having reached phase 2/3, phase 3 or having received emergency authorization. These represent only the tip of the iceberg, since many more antibodies have been discovered and represent opportunities either for diagnosis purposes or as drug candidates. The main problem facing laboratories willing to develop such antibodies is the huge task of analyzing them and choosing the best candidate for exhaustive experimental validation. In this work we show how artificial intelligence-based methods can help in analyzing large sets of antibodies in order to determine in a few hours the best candidates in few hours.