Promiscuity and Specificity of Eukaryotic Glycosyltransferases
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

CRISPR Screening of Porcine Sgrna Library Identifies Host Factors
ARTICLE https://doi.org/10.1038/s41467-020-18936-1 OPEN CRISPR screening of porcine sgRNA library identifies host factors associated with Japanese encephalitis virus replication Changzhi Zhao1,5, Hailong Liu1,5, Tianhe Xiao1,5, Zichang Wang1, Xiongwei Nie1, Xinyun Li1,2, Ping Qian2,3, Liuxing Qin3, Xiaosong Han1, Jinfu Zhang1, Jinxue Ruan1, Mengjin Zhu1,2, Yi-Liang Miao 1,2, Bo Zuo1,2, ✉ ✉ Kui Yang4, Shengsong Xie 1,2 & Shuhong Zhao 1,2 1234567890():,; Japanese encephalitis virus (JEV) is a mosquito-borne zoonotic flavivirus that causes ence- phalitis and reproductive disorders in mammalian species. However, the host factors critical for its entry, replication, and assembly are poorly understood. Here, we design a porcine genome-scale CRISPR/Cas9 knockout (PigGeCKO) library containing 85,674 single guide RNAs targeting 17,743 protein-coding genes, 11,053 long ncRNAs, and 551 microRNAs. Subsequently, we use the PigGeCKO library to identify key host factors facilitating JEV infection in porcine cells. Several previously unreported genes required for JEV infection are highly enriched post-JEV selection. We conduct follow-up studies to verify the dependency of JEV on these genes, and identify functional contributions for six of the many candidate JEV- related host genes, including EMC3 and CALR. Additionally, we identify that four genes associated with heparan sulfate proteoglycans (HSPGs) metabolism, specifically those responsible for HSPGs sulfurylation, facilitate JEV entry into porcine cells. Thus, beyond our development of the largest CRISPR-based functional genomic screening platform for pig research to date, this study identifies multiple potentially vulnerable targets for the devel- opment of medical and breeding technologies to treat and prevent diseases caused by JEV. -

Genome-Wide DNA Methylation Analysis of KRAS Mutant Cell Lines Ben Yi Tew1,5, Joel K
www.nature.com/scientificreports OPEN Genome-wide DNA methylation analysis of KRAS mutant cell lines Ben Yi Tew1,5, Joel K. Durand2,5, Kirsten L. Bryant2, Tikvah K. Hayes2, Sen Peng3, Nhan L. Tran4, Gerald C. Gooden1, David N. Buckley1, Channing J. Der2, Albert S. Baldwin2 ✉ & Bodour Salhia1 ✉ Oncogenic RAS mutations are associated with DNA methylation changes that alter gene expression to drive cancer. Recent studies suggest that DNA methylation changes may be stochastic in nature, while other groups propose distinct signaling pathways responsible for aberrant methylation. Better understanding of DNA methylation events associated with oncogenic KRAS expression could enhance therapeutic approaches. Here we analyzed the basal CpG methylation of 11 KRAS-mutant and dependent pancreatic cancer cell lines and observed strikingly similar methylation patterns. KRAS knockdown resulted in unique methylation changes with limited overlap between each cell line. In KRAS-mutant Pa16C pancreatic cancer cells, while KRAS knockdown resulted in over 8,000 diferentially methylated (DM) CpGs, treatment with the ERK1/2-selective inhibitor SCH772984 showed less than 40 DM CpGs, suggesting that ERK is not a broadly active driver of KRAS-associated DNA methylation. KRAS G12V overexpression in an isogenic lung model reveals >50,600 DM CpGs compared to non-transformed controls. In lung and pancreatic cells, gene ontology analyses of DM promoters show an enrichment for genes involved in diferentiation and development. Taken all together, KRAS-mediated DNA methylation are stochastic and independent of canonical downstream efector signaling. These epigenetically altered genes associated with KRAS expression could represent potential therapeutic targets in KRAS-driven cancer. Activating KRAS mutations can be found in nearly 25 percent of all cancers1. -

NICU Gene List Generator.Xlsx
Neonatal Crisis Sequencing Panel Gene List Genes: A2ML1 - B3GLCT A2ML1 ADAMTS9 ALG1 ARHGEF15 AAAS ADAMTSL2 ALG11 ARHGEF9 AARS1 ADAR ALG12 ARID1A AARS2 ADARB1 ALG13 ARID1B ABAT ADCY6 ALG14 ARID2 ABCA12 ADD3 ALG2 ARL13B ABCA3 ADGRG1 ALG3 ARL6 ABCA4 ADGRV1 ALG6 ARMC9 ABCB11 ADK ALG8 ARPC1B ABCB4 ADNP ALG9 ARSA ABCC6 ADPRS ALK ARSL ABCC8 ADSL ALMS1 ARX ABCC9 AEBP1 ALOX12B ASAH1 ABCD1 AFF3 ALOXE3 ASCC1 ABCD3 AFF4 ALPK3 ASH1L ABCD4 AFG3L2 ALPL ASL ABHD5 AGA ALS2 ASNS ACAD8 AGK ALX3 ASPA ACAD9 AGL ALX4 ASPM ACADM AGPS AMELX ASS1 ACADS AGRN AMER1 ASXL1 ACADSB AGT AMH ASXL3 ACADVL AGTPBP1 AMHR2 ATAD1 ACAN AGTR1 AMN ATL1 ACAT1 AGXT AMPD2 ATM ACE AHCY AMT ATP1A1 ACO2 AHDC1 ANK1 ATP1A2 ACOX1 AHI1 ANK2 ATP1A3 ACP5 AIFM1 ANKH ATP2A1 ACSF3 AIMP1 ANKLE2 ATP5F1A ACTA1 AIMP2 ANKRD11 ATP5F1D ACTA2 AIRE ANKRD26 ATP5F1E ACTB AKAP9 ANTXR2 ATP6V0A2 ACTC1 AKR1D1 AP1S2 ATP6V1B1 ACTG1 AKT2 AP2S1 ATP7A ACTG2 AKT3 AP3B1 ATP8A2 ACTL6B ALAS2 AP3B2 ATP8B1 ACTN1 ALB AP4B1 ATPAF2 ACTN2 ALDH18A1 AP4M1 ATR ACTN4 ALDH1A3 AP4S1 ATRX ACVR1 ALDH3A2 APC AUH ACVRL1 ALDH4A1 APTX AVPR2 ACY1 ALDH5A1 AR B3GALNT2 ADA ALDH6A1 ARFGEF2 B3GALT6 ADAMTS13 ALDH7A1 ARG1 B3GAT3 ADAMTS2 ALDOB ARHGAP31 B3GLCT Updated: 03/15/2021; v.3.6 1 Neonatal Crisis Sequencing Panel Gene List Genes: B4GALT1 - COL11A2 B4GALT1 C1QBP CD3G CHKB B4GALT7 C3 CD40LG CHMP1A B4GAT1 CA2 CD59 CHRNA1 B9D1 CA5A CD70 CHRNB1 B9D2 CACNA1A CD96 CHRND BAAT CACNA1C CDAN1 CHRNE BBIP1 CACNA1D CDC42 CHRNG BBS1 CACNA1E CDH1 CHST14 BBS10 CACNA1F CDH2 CHST3 BBS12 CACNA1G CDK10 CHUK BBS2 CACNA2D2 CDK13 CILK1 BBS4 CACNB2 CDK5RAP2 -

Fucosyltransferase Genes on Porcine Chromosome 6Q11 Are Closely Linked to the Blood Group Inhibitor (S) and Escherichia Coli F18 Receptor (ECF18R) Loci
Mammalian Genome 8, 736–741 (1997). © Springer-Verlag New York Inc. 1997 Two a(1,2) fucosyltransferase genes on porcine Chromosome 6q11 are closely linked to the blood group inhibitor (S) and Escherichia coli F18 receptor (ECF18R) loci E. Meijerink,1 R. Fries,1,*P.Vo¨geli,1 J. Masabanda,1 G. Wigger,1 C. Stricker,1 S. Neuenschwander,1 H.U. Bertschinger,2 G. Stranzinger1 1Institute of Animal Science, Swiss Federal Institute of Technology, ETH-Zentrum, CH-8092 Zurich, Switzerland 2Institute of Veterinary Bacteriology, University of Zurich, CH 8057 Zurich, Switzerland Received: 17 February 1997 / Accepted: 30 May 1997 Abstract. The Escherichia coli F18 receptor locus (ECF18R) has fimbriae F107, has been shown to be genetically controlled by the been genetically mapped to the halothane linkage group on porcine host and is inherited as a dominant trait (Bertschinger et al. 1993) Chromosome (Chr) 6. In an attempt to obtain candidate genes for with B being the susceptibility allele and b the resistance allele. this locus, we isolated 5 cosmids containing the a(1,2)fucosyl- The genetic locus for this E. coli F18 receptor (ECF18R) has been transferase genes FUT1, FUT2, and the pseudogene FUT2P from mapped to porcine Chr 6 (SSC6), based on its close linkage to the a porcine genomic library. Mapping by fluorescence in situ hy- S locus and other loci of the halothane (HAL) linkage group (Vo¨- bridization placed all these clones in band q11 of porcine Chr 6 geli et al. 1996). The epistatic S locus suppresses the phenotypic (SSC6q11). Sequence analysis of the cosmids resulted in the char- expression of the A-0 blood group system when being SsSs (Vo¨geli acterization of an open reading frame (ORF), 1098 bp in length, et al. -

(12) Patent Application Publication (10) Pub. No.: US 2003/0082511 A1 Brown Et Al
US 20030082511A1 (19) United States (12) Patent Application Publication (10) Pub. No.: US 2003/0082511 A1 Brown et al. (43) Pub. Date: May 1, 2003 (54) IDENTIFICATION OF MODULATORY Publication Classification MOLECULES USING INDUCIBLE PROMOTERS (51) Int. Cl." ............................... C12O 1/00; C12O 1/68 (52) U.S. Cl. ..................................................... 435/4; 435/6 (76) Inventors: Steven J. Brown, San Diego, CA (US); Damien J. Dunnington, San Diego, CA (US); Imran Clark, San Diego, CA (57) ABSTRACT (US) Correspondence Address: Methods for identifying an ion channel modulator, a target David B. Waller & Associates membrane receptor modulator molecule, and other modula 5677 Oberlin Drive tory molecules are disclosed, as well as cells and vectors for Suit 214 use in those methods. A polynucleotide encoding target is San Diego, CA 92121 (US) provided in a cell under control of an inducible promoter, and candidate modulatory molecules are contacted with the (21) Appl. No.: 09/965,201 cell after induction of the promoter to ascertain whether a change in a measurable physiological parameter occurs as a (22) Filed: Sep. 25, 2001 result of the candidate modulatory molecule. Patent Application Publication May 1, 2003 Sheet 1 of 8 US 2003/0082511 A1 KCNC1 cDNA F.G. 1 Patent Application Publication May 1, 2003 Sheet 2 of 8 US 2003/0082511 A1 49 - -9 G C EH H EH N t R M h so as se W M M MP N FIG.2 Patent Application Publication May 1, 2003 Sheet 3 of 8 US 2003/0082511 A1 FG. 3 Patent Application Publication May 1, 2003 Sheet 4 of 8 US 2003/0082511 A1 KCNC1 ITREXCHO KC 150 mM KC 2000000 so 100 mM induced Uninduced Steady state O 100 200 300 400 500 600 700 Time (seconds) FIG. -

SUPPLEMENTARY MATERIAL Effect of Next
SUPPLEMENTARY MATERIAL Effect of Next-Generation Exome Sequencing Depth for Discovery of Diagnostic Variants KKyung Kim1,2,3†, Moon-Woo Seong4†, Won-Hyong Chung3, Sung Sup Park4, Sangseob Leem1, Won Park5,6, Jihyun Kim1,2, KiYoung Lee1,2*‡, Rae Woong Park1,2* and Namshin Kim5,6** 1Department of Biomedical Informatics, Ajou University School of Medicine, Suwon 443-749, Korea 2Department of Biomedical Science, Graduate School, Ajou University, Suwon 443-749, Korea, 3Korean Bioinformation Center, Korea Research Institute of Bioscience and Biotechnology, Daejeon 305-806, Korea, 4Department of Laboratory Medicine, Seoul National University Hospital College of Medicine, Seoul 110-799, Korea, 5Department of Functional Genomics, Korea University of Science and Technology, Daejeon 305-806, Korea, 6Epigenomics Research Center, Genome Institute, Korea Research Institute of Bioscience and Biotechnology, Daejeon 305-806, Korea http//www. genominfo.org/src/sm/gni-13-31-s001.pdf Supplementary Table 1. List of diagnostic genes Gene Symbol Description Associated diseases ABCB11 ATP-binding cassette, sub-family B (MDR/TAP), member 11 Intrahepatic cholestasis ABCD1 ATP-binding cassette, sub-family D (ALD), member 1 Adrenoleukodystrophy ACVR1 Activin A receptor, type I Fibrodysplasia ossificans progressiva AGL Amylo-alpha-1, 6-glucosidase, 4-alpha-glucanotransferase Glycogen storage disease ALB Albumin Analbuminaemia APC Adenomatous polyposis coli Adenomatous polyposis coli APOE Apolipoprotein E Apolipoprotein E deficiency AR Androgen receptor Androgen insensitivity -
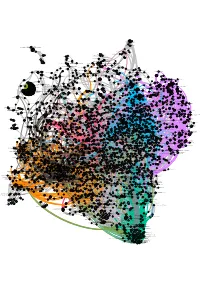
Mitochondrial ABC Transporters Vxpx Cargo-Targeting to Cilium Alpha
PA2GF PA2G5PA2GD PA21BPA2GEPA2GXPA2G3 Acyl chainAcyl chainremodelling remodelling of PI of PG Acyl chain remodelling of PS PA24C AcylAcyl chain chain remodelling remodelling of PCof PE GABA A receptor activation Neurotransmitter receptors and postsynaptic signal transmission GBRA2 GBRB3GBRA6GBRA5 PA2GA GBRA1GBRG2GBRB2GBRB1 GBRT GBRG3GBRA4GBRA3 PA24B Ligand-gated ion channel transport Acyl-CoA:dihydroxyacetonephosphateacyltransferase PPBI Hydrolysis of LPC Hydrolysis ofSynthesis LPE of PA LA Post-translational modification: synthesis of GPI-anchored proteins Catecholamine biosynthesis Plasmalogen biosynthesis GABAGBRR3 AGBRR2 (rho) receptor activation RNA Polymerase III Transcription Termination GBRR1 Creatine metabolism RNA Polymerase III AbortivePNMT AndDDC Retractive Initiation ReuptakeS6A11 of GABA PPBT DigestionPPBN S6A12SC6A7 Interaction between L1 and Ankyrins GLRA1 SC6A1 S6A13 SerotoninSodium/myo-inositol andInositol melatonin transporters cotransporter biosynthesis 2 GLRA2 KCRM FOLR2 KCNQ2 5HT3E5HT3D SC6A3 Astrocytic Glutamate-Glutamine Uptake And Metabolism 5HT3B EAA1 5HT3A5HT3C Na+/Cl- dependent neurotransmitter transportersDopamine clearance from the synaptic cleft SCN8AEAA2 Methylation of TransportINMTMeSeH forof inorganicexcretionSCNAASCN3ASCN2A cations/anions and amino acids/oligopeptidesGlycosyltransferase-likeO-linked glycosylation protein LARGE1 SC6A5SC6A9S6A15 SCN9ASCN4ASCN1ASCNBA SC6A6 SCN7ASCN5A Insulin effects increased synthesis of Xylulose-5-Phosphate SC6A2 TPH1 Potassium transport channels MOT2 Glutamine synthetaseCAC1I -

A-Transferase, 338 ABO Blood Group System, 330 and Cloning, 338
Index A-transferase, 338 ALG9,165 ABO blood group system, 330 ALGlO,168 and cloning, 338 genetic basis of, 22 B-transferase, 338 activated oligosaccharides bacterial S-Iayers in glycopeptide synthesis, 466, 470 glycosylation in, 439 2-aminoethylphosphonate bacterial toxins in invertebrate glycoproteins, 420 and glycosylation, 413 anomer, 5 biological macromolecules formal defmition of, 6 the four groups of, 2 Hudson definition of, 6 bird's nest soup, 16 anomeric carbon blood group antigens, 22 in synthesis, 459 and cancer, 23 anomeric effect, 8 prognostic value of, 23 anomeric oxygen exchange reactions, blood typing, 22 460,461 ABO system of, 22 anomeric oxygen-retaining reactions, 460, Lewis system of, 22 464 Bombay blood group, 331, 336, 348 antennae Bombay phenotype, 22 definition of, 10 branch specificity, 93 J3-arabinofuranose in plants, 418 caeruloplasmin, 23 asialoglycoprotein receptor, 494 calnexin, 54, 182 asparagine-linked glycosylation genes calreticulin, 54, 182, 188 ALGI, 158 capillary electrophoresis, 457 ALG2,161 carbohydrate metabolism, 1, 17 ALG3,165 Carbohydrate-Deficient Glycoprotein ALG5,167 Syndrome (COGS) Type 1, 152 ALG6,168 Carbohydrate-Deficient Glycoprotein ALG7,155 Syndrome (COGS) Type II, 226 ALGB,168 500 Index Carbohydrates; See a/so oligosaccharides deoxymannoj irimycin, 191 as molecules with key biological deoxynojirimycin, 184 functions, 3 dietary sugars, 1 biology of, 2 disaccharides covalent attachment to protein, 17 structural determination of, 17 definition of, 4 DNA,1 in bacterial and viral infection, -

Development and Validation of a Protein-Based Risk Score for Cardiovascular Outcomes Among Patients with Stable Coronary Heart Disease
Supplementary Online Content Ganz P, Heidecker B, Hveem K, et al. Development and validation of a protein-based risk score for cardiovascular outcomes among patients with stable coronary heart disease. JAMA. doi: 10.1001/jama.2016.5951 eTable 1. List of 1130 Proteins Measured by Somalogic’s Modified Aptamer-Based Proteomic Assay eTable 2. Coefficients for Weibull Recalibration Model Applied to 9-Protein Model eFigure 1. Median Protein Levels in Derivation and Validation Cohort eTable 3. Coefficients for the Recalibration Model Applied to Refit Framingham eFigure 2. Calibration Plots for the Refit Framingham Model eTable 4. List of 200 Proteins Associated With the Risk of MI, Stroke, Heart Failure, and Death eFigure 3. Hazard Ratios of Lasso Selected Proteins for Primary End Point of MI, Stroke, Heart Failure, and Death eFigure 4. 9-Protein Prognostic Model Hazard Ratios Adjusted for Framingham Variables eFigure 5. 9-Protein Risk Scores by Event Type This supplementary material has been provided by the authors to give readers additional information about their work. Downloaded From: https://jamanetwork.com/ on 10/02/2021 Supplemental Material Table of Contents 1 Study Design and Data Processing ......................................................................................................... 3 2 Table of 1130 Proteins Measured .......................................................................................................... 4 3 Variable Selection and Statistical Modeling ........................................................................................ -

CDG and Immune Response: from Bedside to Bench and Back Authors
CDG and immune response: From bedside to bench and back 1,2,3 1,2,3,* 2,3 1,2 Authors: Carlota Pascoal , Rita Francisco , Tiago Ferro , Vanessa dos Reis Ferreira , Jaak Jaeken2,4, Paula A. Videira1,2,3 *The authors equally contributed to this work. 1 Portuguese Association for CDG, Lisboa, Portugal 2 CDG & Allies – Professionals and Patient Associations International Network (CDG & Allies – PPAIN), Caparica, Portugal 3 UCIBIO, Departamento Ciências da Vida, Faculdade de Ciências e Tecnologia, Universidade NOVA de Lisboa, 2829-516 Caparica, Portugal 4 Center for Metabolic Diseases, UZ and KU Leuven, Leuven, Belgium Word count: 7478 Number of figures: 2 Number of tables: 3 This article has been accepted for publication and undergone full peer review but has not been through the copyediting, typesetting, pagination and proofreading process which may lead to differences between this version and the Version of Record. Please cite this article as doi: 10.1002/jimd.12126 This article is protected by copyright. All rights reserved. Abstract Glycosylation is an essential biological process that adds structural and functional diversity to cells and molecules, participating in physiological processes such as immunity. The immune response is driven and modulated by protein-attached glycans that mediate cell-cell interactions, pathogen recognition and cell activation. Therefore, abnormal glycosylation can be associated with deranged immune responses. Within human diseases presenting immunological defects are Congenital Disorders of Glycosylation (CDG), a family of around 130 rare and complex genetic diseases. In this review, we have identified 23 CDG with immunological involvement, characterised by an increased propensity to – often life-threatening – infection. -

Viruses Like Sugars: How to Assess Glycan Involvement in Viral Attachment
microorganisms Review Viruses Like Sugars: How to Assess Glycan Involvement in Viral Attachment Gregory Mathez and Valeria Cagno * Institute of Microbiology, Lausanne University Hospital, University of Lausanne, 1011 Lausanne, Switzerland; [email protected] * Correspondence: [email protected] Abstract: The first step of viral infection requires interaction with the host cell. Before finding the specific receptor that triggers entry, the majority of viruses interact with the glycocalyx. Identifying the carbohydrates that are specifically recognized by different viruses is important both for assessing the cellular tropism and for identifying new antiviral targets. Advances in the tools available for studying glycan–protein interactions have made it possible to identify them more rapidly; however, it is important to recognize the limitations of these methods in order to draw relevant conclusions. Here, we review different techniques: genetic screening, glycan arrays, enzymatic and pharmacological approaches, and surface plasmon resonance. We then detail the glycan interactions of enterovirus D68 and severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), highlighting the aspects that need further clarification. Keywords: attachment receptor; viruses; glycan; sialic acid; heparan sulfate; HBGA; SARS-CoV-2; EV-D68 Citation: Mathez, G.; Cagno, V. Viruses Like Sugars: How to Assess 1. Introduction Glycan Involvement in Viral This review focuses on methods for assessing the involvement of carbohydrates in Attachment. Microorganisms 2021, 9, viral attachment and entry into the host cell. Viruses often bind to entry receptors that are 1238. https://doi.org/10.3390/ not abundant on the cell surface; to increase their chances of finding them, they initially microorganisms9061238 bind to attachment receptors comprising carbohydrates that are more widely expressed. -

Supplementary Materials and Tables a and B
SUPPLEMENTARY MATERIAL 1 Table A. Main characteristics of the subset of 23 AML patients studied by high-density arrays (subset A) WBC BM blasts MYST3- MLL Age/Gender WHO / FAB subtype Karyotype FLT3-ITD NPM status (x109/L) (%) CREBBP status 1 51 / F M4 NA 21 78 + - G A 2 28 / M M4 t(8;16)(p11;p13) 8 92 + - G G 3 53 / F M4 t(8;16)(p11;p13) 27 96 + NA G NA 4 24 / M PML-RARα / M3 t(15;17) 5 90 - - G G 5 52 / M PML-RARα / M3 t(15;17) 1.5 75 - - G G 6 31 / F PML-RARα / M3 t(15;17) 3.2 89 - - G G 7 23 / M RUNX1-RUNX1T1 / M2 t(8;21) 38 34 - + ND G 8 52 / M RUNX1-RUNX1T1 / M2 t(8;21) 8 68 - - ND G 9 40 / M RUNX1-RUNX1T1 / M2 t(8;21) 5.1 54 - - ND G 10 63 / M CBFβ-MYH11 / M4 inv(16) 297 80 - - ND G 11 63 / M CBFβ-MYH11 / M4 inv(16) 7 74 - - ND G 12 59 / M CBFβ-MYH11 / M0 t(16;16) 108 94 - - ND G 13 41 / F MLLT3-MLL / M5 t(9;11) 51 90 - + G R 14 38 / F M5 46, XX 36 79 - + G G 15 76 / M M4 46 XY, der(10) 21 90 - - G NA 16 59 / M M4 NA 29 59 - - M G 17 26 / M M5 46, XY 295 92 - + G G 18 62 / F M5 NA 67 88 - + M A 19 47 / F M5 del(11q23) 17 78 - + M G 20 50 / F M5 46, XX 61 59 - + M G 21 28 / F M5 46, XX 132 90 - + G G 22 30 / F AML-MD / M5 46, XX 6 79 - + M G 23 64 / M AML-MD / M1 46, XY 17 83 - + M G WBC: white blood cell.