Manipulation of the Microbiome and Its Impact on Functional Recovery Following Ischemic Stroke Michal Jandzinski [email protected]
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Noninvasive Evaluation of Myocardial Ischemia and Left Ventricular Function
Linköping University Medical Dissertations No. 1109 Noninvasive Evaluation of Myocardial Ischemia and Left Ventricular Function Eva Maret Division of Cardiovascular Medicine Department of Medical and Health Sciences Linköping University, Sweden Linköping 2009 Eva Maret, 2009 Cover picture/illustration: William Björklund Published articles have been reprinted with the permission of the copyright holder. Printed in Sweden by LiU-Tryck, Linköping, Sweden, 2009 ISBN 978-91-7393-675-0 ISSN 0345-0082 To Martin and John! Somewhere over the rainbow Way up high, There's a land that I heard of Once in a lullaby. Somewhere over the rainbow Skies are blue, And the dreams that you dare to dream Really do come true. From “The Wizard of OZ”, music by Harold Arlen and lyrics by E.Y. Harburg CONTENTS POPULÄRVETENSKAPLIG SAMMANFATTNING ................................................................ 7 LIST OF PUBLICATIONS .......................................................................................................... 9 INTRODUCTION ....................................................................................................................... 13 Coronary Artery Disease ........................................................................................................ 13 Coronary flow, resistance and flow reserve ........................................................................... 14 Coronary atherosclerosis and myocardial infarction .............................................................. 15 Left ventricular function -

Chapter 1: Stroke and Neuroprotection 1 – 21
DEVELOPMENT OF NOVEL THERAPEUTICS FOR STROKE: PRECLINICAL INVESTIGATIONS OF OSTEOPONTIN AND 3-IODOTHYRONAMINE By Kristian Paul Doyle A DISSERTATION Presented to the Department of Molecular Microbiology & Immunology at the Oregon Health & Science University in partial fulfillment of the requirements for the degree of Doctor of Philosophy 1 CONTENTS List of Figures v List of Tables ix Acknowledgements x Preface xi Abstract xii List of Abbreviations xv Chapter 1: Stroke and Neuroprotection 1 – 21 1.1 Introduction 2 1.2 Brief History of Stroke 2 1.3 Stroke Pathophysiology 4 1.4 Neuroprotection 17 1.5 Ischemic Preconditioning 19 1.6 Research Goal 21 Chapter 2: Osteopontin 22-86 2.1 An Introduction to OPN 23 2.2 The Structure of OPN 23 2.3 OPN, Integrins and Survival Signaling 25 2.4 OPN and Ischemic Injury 27 2.5 Preclinical Development of OPN 33 2.6 Optimizing Delivery 33 2 2.7 Improving the Potency of OPN 36 2.8 Identifying the Regions of OPN required for Neuroprotection 36 2.9 Hypothesis 37 2.10 Research Design 38 2.11 OPN has neuroprotective capability in vivo and in vitro 40 2.12 The mechanism of neuroprotection by OPN 51 2.13 OPN can be delivered to the brain by intranasal administration 56 2.14 Enhancing the neuroprotective capability of OPN 60 2.15 Peptides based on the N and C terminal fragment of thrombin cleaved OPN are neuroprotective 65 2.16 The C terminal peptide requires phosphorylation to be neuroprotective while the N terminal peptide does not require phosphorylation 70 2.17 Dose response and time window of NT 124-153 71 2.18 -

College Catalog 2011–2012
1 College Catalog 2011–2012 - BOSTON | WORCESTER | MANCHESTER, NH This catalog is intended to provide working guidelines and descriptions of the general and academic policies of the College applicable to students. It is not intended and cannot be Massachusetts College of Pharmacy and Health Sciences construed as a contract or guaranty of any kind, express or implied, and the College may 179 Longwood Avenue, Boston, Massachusetts 02115 change, delete or add to these guidelines unilaterally in its sole discretion and without notice. The College also reserves the right to determine the applicability of any policy to a particular Telephone 617.732.2800; students outside Massachusetts and within the continental United situation or set of circumstances and to depart from the guidelines contained herein in States may call toll free 1.800.225.5506. Non-Discrimination Policy a given case. This catalog supersedes any previous catalog, policies or practices relating It is the policy and commitment of Massachusetts College of Pharmacy and Health Sciences 2 to students. It is the responsibility of the students to know and understand the College’s 3 not to discriminate on the basis of race, religion, color, age, sexual orientation, sex, sexual policies. The College may from time to time acquire or develop new programs, or expand its identity, disability, veteran status, marital status or national origin in its educational pro- offerings in other locations, including distance learning programs, and the guidelines in this grams, activities, admissions or employment policies and to actively comply with the require- ments of Federal Executive Orders 11246 and 11375 as amended; the Civil Rights Act of catalog shall apply to all such programs and locations. -

Ischemia and Reperfusion Injury: When Cells Almost Die
Peer Reviewed CE Article #1 Ischemia and Reperfusion Injury: When Cells Almost Die Amy N. Breton, CVT, VTS (ECC) Veterinary Emergency and Specialty Center of New England Waltham, Massachusetts schemia is defined as inadequate blood supply to a part ing energy for metabolism within cells.3 One of the fastest of the body, usually caused by partial or total block- ways that ATP is produced is by oxidative phosphorylation, age of an artery. Reperfusion injury occurs when tissue which, as the name implies, requires oxygen.3 Despite the I 3 perfusion and oxygenation are restored to an area that has importance of ATP, cells do not stockpile ATP. They only been affected by an ischemic event. make what they need for a particular time. When ischemia Ischemia/reperfusion (I/R) injury is a complex cascade occurs, oxygenation of cells ceases, resulting in anaerobic of events resulting in devastating effects on the body, ATP production, which is less efficient.3 sometimes including death. Despite more than 70 years of When oxygen becomes unavailable, cells begin anaero- research, I/R injury is not fully understood. Events such as bic glycolysis. This process can be a lifesaving way for cells gastric dilatation–volvulus (GDV), mesenteric torsion, or to obtain energy; however, it is extremely wasteful. During strangulation of a limb can lead to I/R injury. It is important anaerobic glycolysis, pyruvic acid and hydrogen atoms for all veterinary personnel to understand I/R injury so that combine with nicotinamide adenine dinucleotide (NAD) treatment and prevention can begin as early as possible. to form NADH and H+.3 If the buildup of NADH and H+ within cells becomes too great, the anaerobic process stops, The Ischemic Cascade terminating energy production.3 However, NADH and H+ The chain of events involved in I/R injury can be broken combine to form lactic acid, which diffuses from cells rap- down into the ischemic cascade (BOX 1) and reperfusion idly so that the process can continue.3 Although this is not injury (BOX 2). -

2020 Vascular Neurology CERT Content Specifications
SUBSPECIALTY CERTIFICATION EXAMINATION IN VASCULAR NEUROLOGY 2020 Content Blueprint (December 16, 2019) Number of questions: 200 questions Percent 01. Basic science aspects of vascular neurology 4-6% 02. Risk factors and epidemiology 8-12% 03. Clinical features of cerebrovascular diseases 8-12% 04. Evaluation of the patient with cerebrovascular disease 13-17% 05. Causes of stroke 18-22% 06. Complications of stroke 4-6% 07. Treatment of patients with stroke 28-32% 08. Recovery, regenerative approaches, and rehabilitation 4-6% TOTAL 100% Note: A more detailed content outline is shown below 2020 ABPN Content Specifications Page 1 of 15 Posted January 6, 2020 Certification in Vascular Neurology SUBSPECIALTY CERTIFICATION EXAMINATION IN VASCULAR NEUROLOGY 2020 Content Outline Content Areas 01. Basic science aspects of vascular neurology A. Vascular neuroanatomy 1. Extracranial arterial anatomy 2. Intracranial arterial anatomy 3. Collaterals 4. Alterations of vascular anatomy 5. Venous anatomy 6. Spinal cord vascular anatomy 7. Specific vascular-brain anatomic correlations 8. End vessel syndromes B. Stroke pathophysiology 1. Cerebral blood flow a. Vascular smooth muscle control b. Vasodilation and vasoconstriction c. Autoregulation d. Vasospasm e. Rheology f. Blood flow in stroke 2. Blood-brain barrier in stroke 3. Coagulation cascade a. Clotting factors b. Platelet function c. Endothelium function d. Biochemical factors 4. Metabolic and cellular consequences of ischemia a. Ischemic cascade b. Reperfusion changes c. Electrophysiology d. Gene regulation 5. Inflammation and stroke 6. Brain edema and increased ICP a. Secondary effects 2020 ABPN Content Specifications Page 2 of 15 Posted January 6, 2020 Certification in Vascular Neurology 7. Restoration and recovery following stroke 8. -

Reduced Susceptibility to Ischemic Brain Injury and N-Methyl-D-Aspartate-Mediated Neurotoxicity in Cyclooxygenase-2-Deficient Mice
Reduced susceptibility to ischemic brain injury and N-methyl-D-aspartate-mediated neurotoxicity in cyclooxygenase-2-deficient mice Costantino Iadecola*†, Kiyoshi Niwa*, Shigeru Nogawa*, Xueren Zhao*, Masao Nagayama*, Eiichi Araki*, Scott Morham‡, and M. Elizabeth Ross* *Center for Clinical and Molecular Neurobiology, Department of Neurology, University of Minnesota Medical School, Minneapolis, MN 55455; and ‡Myriad Genetics, Salt Lake City, UT 84108 Communicated by Philip Needleman, Monsanto Company, St. Louis, MO, November 21, 2000 (received for review July 27, 2000) Cyclooxygenase-2 (COX-2), a prostanoid-synthesizing enzyme that 29). In rodents as in humans, cerebral ischemia up-regulates contributes to the toxicity associated with inflammation, has COX-2 in neurons, blood vessels, and inflammatory cells infil- recently emerged as a promising therapeutic target for several trating the injured brain (27, 28, 30). However, experimental illnesses, ranging from osteoarthritis to Alzheimer’s disease. Al- evidence linking COX-2 to the mechanisms of brain injury though COX-2 has also been linked to ischemic stroke, its role in the associated with these conditions is lacking. For example, al- mechanisms of ischemic brain injury remains controversial. We though it is well established that cerebral ischemia increases demonstrate that COX-2-deficient mice have a significant reduc- COX-2 expression in the damaged brain, studies using COX tion in the brain injury produced by occlusion of the middle inhibitors in models of focal cerebral ischemia have yielded cerebral artery. The protection can be attributed to attenuation conflicting results (28, 31, 32). Furthermore, studies using of glutamate neurotoxicity, a critical factor in the initiation of COX-2 inhibitors cannot exclude effects unrelated to COX-2, ischemic brain injury, and to abrogation of the deleterious effects of postischemic inflammation, a process contributing to the sec- such as modification of gene expression or activation of perox- ondary progression of the damage. -

Impact of an Immune Modulator Fingolimod on Acute Ischemic Stroke
Impact of an immune modulator fingolimod on acute ischemic stroke Ying Fua,b, Ningnannan Zhangc, Li Rena, Yaping Yana, Na Suna, Yu-Jing Lia, Wei Hanc, Rong Xuea, Qiang Liua,b, Junwei Haoa, Chunshui Yuc, and Fu-Dong Shia,b,d,1 Departments of aNeurology and cRadiology, Tianjin Neurological Institute, Tianjin Medical University General Hospital, Tianjin, 300070, China; bDepartment of Neurology, Barrow Neurological Institute, St. Joseph’s Hospital and Medical Center, Phoenix, AZ 85013; and dDepartment of Immunology, Tianjin Medical University, Tianjin, 300070, China Edited by Solomon H. Snyder, The Johns Hopkins University School of Medicine, Baltimore, MD, and approved November 5, 2014 (received for review August 21, 2014) Peripheral lymphocytes entering brain ischemic regions orches- brain (7, 8). Cell types most frequently known to enter the brain + + trate inflammatory responses, catalyze tissue death, and worsen during AIS belong to the CD4 T-cell, CD8 T-cell, neutrophil, clinical outcomes of acute ischemic stroke (AIS) in preclinical and macrophage, and natural killer (NK) cell subpopulations. studies. However, it is not known whether modulating brain Such cells, as infiltrate within the peri-infarcted areas of brain inflammation can impact the outcome of patients with AIS. In this tissues from AIS patients (9, 10), become intimately involved in open-label, evaluator-blinded, parallel-group clinical pilot trial, we all stages of the ischemic cascade (7, 8). Studies in experimental recruited 22 patients matched for clinical and MRI characteristics, stroke have indicated that all these cells contribute to the death with anterior cerebral circulation occlusion and onset of stroke of ischemic neurons by promoting focal inflammatory reactions, that had exceeded 4.5 h, who then received standard manage- direct killing, triggering of antigen-specific immune responses, or ment alone (controls) or standard management plus fingolimod alterations in neuronal excitability (6, 10–12). -

Guidelines for Performance, Interpretation, and Application of Stress Echocardiography in Ischemic Heart Disease: from the American Society of Echocardiography
GUIDELINES AND STANDARDS Guidelines for Performance, Interpretation, and Application of Stress Echocardiography in Ischemic Heart Disease: From the American Society of Echocardiography Patricia A. Pellikka, MD, FASE, Chair, Adelaide Arruda-Olson, MD, PhD, FASE, Farooq A. Chaudhry, MD, FASE,* Ming Hui Chen, MD, MMSc, FASE, Jane E. Marshall, RDCS, FASE, Thomas R. Porter, MD, FASE, and Stephen G. Sawada, MD, Rochester, Minnesota; New York, New York; Boston, Massachusetts; Omaha, Nebraska; Indianapolis, Indiana Keywords: Echocardiography, Stress, Guidelines, Imaging, Ischemic heart disease, Stress test, Pediatrics This document is endorsed by the following ASE International Alliance Partners: Argentine Federation of Cardiology, Argentine Society of Cardiology, ASEAN Society of Echocardiography, Association of Echocardiography and Cardiovascular Imaging of the Interamerican Society of Cardiology, Australasian Sonographers Association, Canadian Society of Echocardiography, Chinese Society of Echocardiography, Cuban Society of Cardiography Echocardiography Section, Department of Cardiovascular Imaging of the Brazilian Society of Cardiology, Indian Academy of Echocardiography, Indian Association of Cardiovascular Thoracic Anaesthesiologists, Indonesian Society of Echocardiography, Iranian Society of Echocardiography, Israeli Working Group on Echocardiography, Italian Association of CardioThoracic and Vascular Anaesthesia and Intensive Care, Japanese Society of Echocardiography, Korean Society of Echocardiography, Mexican Society of Echocardiography -
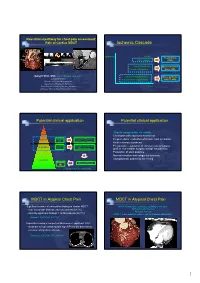
1 Ischemic Cascade
New clinical pathway for chest pain assessment: Role of Cardiac MDCT Ischemic Cascade Ischemia Angina Treadmill test ECG change (TMT) Systolic Dysfunction Stress functional Diastolic Dysfunction (Echo, MRI) Sang Il Choi, M.D. ([email protected]) Transmural Hypoperfusion Stress perfusion Assistant Professor Subendocardial Hypoperfusion (SPECT, MRI) Director of CT and 3D Imaging lab Department of Diagnostic Radiology Seoul National University Bundang Hospital Time College of Medicine Seoul National University Potential clinical application Potential clinical application Sudden Death • Atypical, symptomatic, chest pain Angina • Chest pain with equivocal stress test Symptoms Cardiac Exclusion/Presence • Preprocedural evaluation of chronic total occlusion CT of Clinical Disease • Acute coronary syndrome Cardiac Pre-Clinical Disease • Preoperative evaluation of coronary artery bypass High Risk CT Wall Changes graft or non-cardiac surgery at high risk patients • Evaluation of stent patency • Normal variation and congenital anomaly Low Risk • Asymptomatic patient for screening Prevent Medical Therapy MI Fayad et al, Circulation 2002 MDCT in Atypical Chest Pain MDCT in Atypical Chest Pain • Significant number of noncardiac findings in cardiac MDCT: (M/61) Chest pain: continuous, radiating to the back - new, noncaridac findings: 292/346 patients (58.1%) TMT and Holter: Normal EchoCG: Normal - clinically significant findings: 114/346 patients (22.7%) SPECT: Fixed defect at anterior wall (R/O Breast attenuation) Onuma Y. JACC 2006; 04.071v1. -

Neuroinflammation After Acute Ischemic Stroke: a Volcano Hard to Contain
中国现代神经疾病杂志 年 月第 卷第 期 · 964 · 2013 11 13 11 Chin J Contemp Neurol Neurosurg, November 2013, Vol. 13, No. 11 Reviews · · Neuroinflammation after acute ischemic stroke: a volcano hard to contain Monika Mishra, Vishnumurthy Shushrutha Hedna Department of Neurology, College of Medicine, University of Florida, Gainesville, Florida, USA Keywords Brain ischemia; Inflammation; Central Abstract nervous system; Immunity, celluar; Many endogenous, exogenous and systemic factors individually can lead Neuroimmunomodulation; Review. to occlusive vessel pathology affecting the neuroendovascular unit 【关键词】 脑缺血; 炎症; 中枢神经 compromising cerebral blood flow. The brain ischemia thus created 系 统 ; 免 疫 ,细胞 ; 神 经 免 疫 调 节 ; irrespective of the pathology has similar endpoint and involves blood 综述 vessels, astrocytes, neurons and surrounding microglia triggering the whole spectrum of neuroinflammation which is an important component Correspondence in the ischemic cascade. The resultant series of neuroinflammatory Vishnumurthy Shushrutha Hedna, Assistant reactions cause depolarization of neuronal cells and activation of pro Professor, Department of Neurology ⁃ inflammatory cellular agents and subsequently cell death. Room L3-100, McKnight Brain Institute Neuroinflammation can be an effect and (or) cause of acute energy failure, 1149 Newell Drive, Gainesville, FL 32611 excitotoxicity, ionic imbalance, channel dysfunctions, and oxidative free Tel: +11 352 273 5550 radicals in the central nervous system. Non restoration of the blood flow Fax: +11 352 273 5575 ⁃ Email: [email protected] within the threshold period can result in an extensive brain damage from activation of deadly latent proteases like matrix metalloprotease, and Received: 11 August, 2013. immediate early genes destroying the neuronal microenvironment and blood brain barrier. -

Role of Herbal Teas in Regulating Cellular Homeostasis and Autophagy and Their Implications in Regulating Overall Health
nutrients Review Role of Herbal Teas in Regulating Cellular Homeostasis and Autophagy and Their Implications in Regulating Overall Health James Michael Brimson 1,2 , Mani Iyer Prasanth 1,2, Dicson Sheeja Malar 1,2, Rajasekharan Sharika 3, Bhagavathi Sundaram Sivamaruthi 4, Periyanaina Kesika 4 , Chaiyavat Chaiyasut 4 , Tewin Tencomnao 1,2,* and Anchalee Prasansuklab 1,5,* 1 Natural Products for Neuroprotection and Anti-Ageing Research Unit, Chulalongkorn University, Bangkok 10330, Thailand; [email protected] (J.M.B.); [email protected] (M.I.P.); [email protected] (D.S.M.) 2 Department of Clinical Chemistry, Faculty of Allied Health Sciences, Chulalongkorn University, Bangkok 10330, Thailand 3 Department of Transfusion Medicine and Clinical Microbiology, Faculty of Allied Health Sciences, Chulalongkorn University, Bangkok 10330, Thailand; [email protected] 4 Innovation Center for Holistic Health, Nutraceuticals, and Cosmeceuticals, Faculty of Pharmacy, Chiang Mai University, Chiang Mai 50200, Thailand; [email protected] (B.S.S.); [email protected] (P.K.); [email protected] (C.C.) 5 College of Public Health Sciences, Chulalongkorn University, Bangkok 10330, Thailand * Correspondence: [email protected] (T.T.); [email protected] (A.P.) Citation: Brimson, J.M.; Prasanth, M.I.; Malar, D.S.; Sharika, R.; Abstract: Tea is one of the most popular and widely consumed beverages worldwide, and possesses Sivamaruthi, B.S.; Kesika, P.; numerous potential health benefits. Herbal teas are well-known to contain an abundance of polyphe- Chaiyasut, C.; Tencomnao, T.; nol antioxidants and other ingredients, thereby implicating protection and treatment against various Prasansuklab, A. Role of Herbal Teas ailments, and maintaining overall health in humans, although their mechanisms of action have not in Regulating Cellular Homeostasis yet been fully identified. -

Novel Treatments for Transient Ischemic Attack and Acute Ischemic Stroke
Novel Treatments for Transient Ischemic Attack and Acute Ischemic Stroke a,b, c,d,e Matthew S. Siket, MD, MSc *, Rhonda Cadena, MD KEYWORDS Acute ischemic stroke Intravenous alteplase Endovascular thrombectomy KEY POINTS Nearly 700,000 people experience a stroke every year and 240,000 experience a transient ischemic attack. Primary prevention is important in stroke prevention and includes controlling hyperten- sion, diabetes, cholesterol, weight, and cardiac risk factors, and getting plenty of physical activity, abstaining from cigarette smoking and alcohol, and eating a nutritious diet. In patients who are not candidates for reperfusion therapy, treatment involves perfusing the ischemic penumbra to prevent worsening of the infarcted region. INTRODUCTION Each year, nearly 700,000 people will suffer an acute ischemic stroke (AIS)1 and an additional 240,000 people will experience a transient ischemic attack (TIA).2 Before the coronavirus disease-19 outbreak, stroke was the fifth leading cause of death in the United States and remains a leading cause of long-term disability. Over the last 25 years, researchers have been searching for novel ways of treating TIA and AIS that are both effective and safe. Although researchers have yet to identify better treat- ments, recent trials have shown better patient selection to extend the time window for some beyond the standard 3-hour window. a Division of Emergency Medicine, Department of Surgery, Larner College of Medicine at the University of Vermont, 111 Colchester Avenue, EC2-216, Burlington,