An Ecological Framework for Modeling the Geography Of
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

COVID-19: Perspective, Patterns and Evolving Strategies
COVID-19: Perspective, Patterns and Evolving strategies Subject Category: Clinical Virology Vinod Nikhra* Department of Medicine, Hindu Rao Hospital & NDMC Medical College, New Delhi, India Submitted: 02 June 2020 | Approved: 06 July 2020 | Published: 09 July 2020 Copyright: © 2020 Nikhra V. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. DOI: https://dx.doi.org/10.29328/ebook1003 ORCID: https://orcid.org/0000-0003-0859-5232 *Corresponding author: Dr. Vinod Nikhra, M.D. Consultant and Faculty, Department of Medicine, Hindu Rao Hospital & NDMC Medical College, New Delhi, India, Tel: 91-9810874937; Email: [email protected]; drvinodnikhra@rediff mail.com Open Access COVID-19: Perspective, Patterns and Evolving strategies Table of Contents - 7 Chapters Sl No Chapters Title Pages The Trans-Zoonotic Virome Interface: Measures to 1 Chapter 1 003-011 Balance, Control and Treat Epidemics Exploring Pathophysiology of COVID-19 Infection: Faux 2 Chapter 2 012-020 Espoir and Dormant Therapeutic Options The Agent and Host Factors in COVID-19: Exploring 3 Chapter 3 021-036 Pathogenesis and Therapeutic Implications Adverse Outcomes for Elderly in COVID-19: Annihilation 4 Chapter 4 037-047 of the Longevity Dream Identifying Patterns in COVID-19: Morbidity, Recovery, 5 Chapter 5 048-058 and the Aftermath The New Revelations: Little-known Facts about COVID-19 6 Chapter 6 059-068 and their Implications Fear, Reaction and Rational Behaviour to COVID-19 in 7 Chapter 7 069-076 Public, Health Professionals and Policy Planners La Confusion: Caring for COVID-19 patients 8 Postscript 077-079 and the raging, engulfi ng and debilitating pandemic 9 Acknowledgement 080-080 *Corresponding HTTPS://WWW.HEIGHPUBS.ORG author: Dr. -

Assembling Evidence for Identifying Reservoirs of Infection
TREE-1806; No. of Pages 10 Review Assembling evidence for identifying reservoirs of infection 1 1,2 1,3 1 4 Mafalda Viana , Rebecca Mancy , Roman Biek , Sarah Cleaveland , Paul C. Cross , 3,5 1 James O. Lloyd-Smith , and Daniel T. Haydon 1 Boyd Orr Centre for Population and Ecosystem Health, Institute of Biodiversity, Animal Health and Comparative Medicine, College of Medical Veterinary and Life Sciences, University of Glasgow, Glasgow G12 8QQ, UK 2 School of Computing Science, University of Glasgow, Glasgow G12 8QQ, UK 3 Fogarty International Center, National Institutes of Health, Bethesda, MD 20892, USA 4 US Geological Survey, Northern Rocky Mountain Science Center 2327, University Way, Suite 2, Bozeman, MT 59715, USA 5 Department of Ecology and Evolutionary Biology, University of California at Los Angeles, Los Angeles, CA 90095, USA Many pathogens persist in multihost systems, making patterns of incidence and prevalence (see Glossary) that the identification of infection reservoirs crucial for result from the connectivity between source and target devising effective interventions. Here, we present a con- populations (black arrows in Figure I in Box 1). We then ceptual framework for classifying patterns of incidence review methods that allow us to identify maintenance or and prevalence, and review recent scientific advances nonmaintenance populations (squares or circles in Figure that allow us to study and manage reservoirs simulta- I, Box 1), how they are connected (arrows in Figure I, Box neously. We argue that interventions can have a crucial 1), and the role that each of these populations has in role in enriching our mechanistic understanding of how maintaining the pathogen (i.e., reservoir capacity). -

Variation in Antimicrobial Susceptibility Among Borrelia Burgdorferi Strains? Emir Hodzic*
BOSNIAN JOURNAL OF BASIC MEDICAL SCIENCES REVIEW WWW.BJBMS.ORG Lyme Borreliosis: is there a preexisting (natural) variation in antimicrobial susceptibility among Borrelia burgdorferi strains? Emir Hodzic* Real-Time PCR Research and Diagnostic Core Facility, School of Veterinary Medicine, University of California at Davis, California, United States of America ABSTRACT The development of antibiotics changed the world of medicine and has saved countless human and animal lives. Bacterial resistance/tolerance to antibiotics have spread silently across the world and has emerged as a major public health concern. The recent emergence of pan-resistant bacteria can overcome virtually any antibiotic and poses a major problem for their successful control. Selection for antibiotic resistance may take place where an antibiotic is present: in the skin, gut, and other tissues of humans and animals and in the environment. Borrelia burgdorferi, the etiological agents of Lyme borreliosis, evades host immunity and establishes persistent infections in its mammalian hosts. The persistent infection poses a challenge to the effective antibiotic treatment, as demonstrated in various animal models. An increasingly heterogeneous sub- population of replicatively attenuated spirochetes arises following treatment, and these persistent antimicrobial tolerant/resistant spirochetes are non-cultivable. The non-cultivable spirochetes resurge in multiple tissues at 12 months after treatment, withB. burgdorferi-specific DNA copy levels nearly equivalent to those found in shame-treated experimental animals. These attenuated spirochetes remain viable, but divide slowly, thereby being tolerant to antibiotics. Despite the continued non-cultivable state, RNA transcription of multiple B. burgdorferi genes was detected in host tissues, spirochetes were acquired by xenodiagnostic ticks, and spirochetal forms could be visualized within ticks and mouse tissues. -

Impact of Babesia Microti Infection on the Initiation and Course Of
Tołkacz et al. Parasites Vectors (2021) 14:132 https://doi.org/10.1186/s13071-021-04638-0 Parasites & Vectors RESEARCH Open Access Impact of Babesia microti infection on the initiation and course of pregnancy in BALB/c mice Katarzyna Tołkacz1,2*, Anna Rodo3, Agnieszka Wdowiarska4, Anna Bajer1† and Małgorzata Bednarska5† Abstract Background: Protozoa in the genus Babesia are transmitted to humans through tick bites and cause babesiosis, a malaria-like illness. Vertical transmission of Babesia spp. has been reported in mammals; however, the exact timing and mechanisms involved are not currently known. The aims of this study were to evaluate the success of vertical transmission of B. microti in female mice infected before pregnancy (mated during the acute or chronic phases of Babesia infection) and that of pregnant mice infected during early and advanced pregnancy; to evaluate the pos- sible infuence of pregnancy on the course of parasite infections (parasitaemia); and to assess pathological changes induced by parasitic infection. Methods: The frst set of experiments involved two groups of female mice infected with B. microti before mating, and inseminated on the 7th day and after the 40th day post infection. A second set of experiments involved female mice infected with B. microti during pregnancy, on the 4th and 12th days of pregnancy. Blood smears and PCR targeting the 559 bp 18S rRNA gene fragment were used for the detection of B. microti. Pathology was assessed histologically. Results: Successful development of pregnancy was recorded only in females mated during the chronic phase of infection. The success of vertical transmission of B. -

MODELING the SPREAD of the 1918 INFLUENZA PANDEMIC in a NEWFOUNDLAND COMMUNITY a Dissertation Presented to the Faculty of the Gr
MODELING THE SPREAD OF THE 1918 INFLUENZA PANDEMIC IN A NEWFOUNDLAND COMMUNITY A Dissertation presented to the Faculty of the Graduate School at the University of Missouri In Partial Fulfillment of the Requirements for the Degree Doctor of Philosophy By JESSICA LEA DIMKA Dr. Lisa Sattenspiel, Dissertation Supervisor MAY 2015 The undersigned, appointed by the dean of the Graduate School, have examined the dissertation entitled MODELING THE SPREAD OF THE 1918 INFLUENZA PANDEMIC IN A NEWFOUNDLAND COMMUNITY Presented by Jessica Lea Dimka A candidate for the degree of Doctor of Philosophy And hereby certify that, in their opinion, it is worthy of acceptance. Professor Lisa Sattenspiel Professor Gregory Blomquist Professor Mary Shenk Professor Enid Schatz ACKNOWLEDGEMENTS This research could not have been completed without the support and guidance of many people who deserve recognition. Dr. Lisa Sattenspiel provided the largest amount of assistance and insight into this project, from initial development through model creation and data analysis to the composition of this manuscript. She has been an excellent mentor over the last seven years. I would also like to extend my gratitude to my committee members – Dr. Greg Blomquist, Dr. Mary Shenk, and Dr. Enid Schatz – for their advice, comments, patience and time. I also would like to thank Dr. Craig Palmer for his insight and support on this project. Additionally, I am grateful to Dr. Allison Kabel, who has provided me with valuable experience, advice and support in my research and education activities while at MU. Many thanks go to the librarians and staff of the Provincial Archives of Newfoundland and Labrador and the Centre for Newfoundland Studies at Memorial University of Newfoundland. -

Prediction and Prevention of the Next Pandemic Zoonosis
Series Zoonoses 3 Prediction and prevention of the next pandemic zoonosis Stephen S Morse, Jonna A K Mazet, Mark Woolhouse, Colin R Parrish, Dennis Carroll, William B Karesh, Carlos Zambrana-Torrelio, W Ian Lipkin, Peter Daszak Lancet 2012; 380: 1956–65 Most pandemics—eg, HIV/AIDS, severe acute respiratory syndrome, pandemic infl uenza—originate in animals, See Comment pages 1883 are caused by viruses, and are driven to emerge by ecological, behavioural, or socioeconomic changes. Despite their and 1884 substantial eff ects on global public health and growing understanding of the process by which they emerge, no This is the third in a Series of pandemic has been predicted before infecting human beings. We review what is known about the pathogens that three papers about zoonoses emerge, the hosts that they originate in, and the factors that drive their emergence. We discuss challenges to their Mailman School of Public control and new eff orts to predict pandemics, target surveillance to the most crucial interfaces, and identify Health (Prof S S Morse PhD), and Center for Infection and prevention strategies. New mathematical modelling, diagnostic, communications, and informatics technologies can Immunity (Prof W I Lipkin MD); identify and report hitherto unknown microbes in other species, and thus new risk assessment approaches are Columbia University, needed to identify microbes most likely to cause human disease. We lay out a series of research and surveillance New York, NY, USA; One Health opportunities and goals that could help to overcome these challenges and move the global pandemic strategy from Institute, School of Veterinary Medicine, University of response to pre-emption. -

Bats Are a Major Natural Reservoir for Hepaciviruses and Pegiviruses
Bats are a major natural reservoir for hepaciviruses and pegiviruses Phenix-Lan Quana,1, Cadhla Firtha, Juliette M. Contea, Simon H. Williamsa, Carlos M. Zambrana-Torreliob, Simon J. Anthonya,b, James A. Ellisonc, Amy T. Gilbertc, Ivan V. Kuzminc,2, Michael Niezgodac, Modupe O. V. Osinubic, Sergio Recuencoc, Wanda Markotterd, Robert F. Breimane, Lems Kalembaf, Jean Malekanif, Kim A. Lindbladeg, Melinda K. Rostalb, Rafael Ojeda-Floresh, Gerardo Suzanh, Lora B. Davisi, Dianna M. Blauj, Albert B. Ogunkoyak, Danilo A. Alvarez Castillol, David Moranl, Sali Ngamm, Dudu Akaiben, Bernard Agwandao, Thomas Briesea, Jonathan H. Epsteinb, Peter Daszakb, Charles E. Rupprechtc,3, Edward C. Holmesp, and W. Ian Lipkina aCenter for Infection and Immunity, Mailman School of Public Health, Columbia University, New York, NY 10032; bEcoHealth Alliance, New York, NY 10001; cPoxvirus and Rabies Branch, Division of High-Consequence Pathogens and Pathology, National Center for Emerging Zoonotic Infectious Diseases, Centers for Disease Control and Prevention, Atlanta, GA 30333; dDepartment of Microbiology and Plant Pathology, University of Pretoria, Pretoria 0002, South Africa; eCenters for Disease Control and Prevention in Kenya, Nairobi, Kenya; fUniversity of Kinshasa, Kinshasa 11, Democratic Republic of the Congo; gCenters for Disease Control and Prevention Guatemala, 01015, Guatemala City, Guatemala; hFacultad de Medicina Veterinaria y Zootecnia, Universidad Nacional Autónoma de México, Ciudad Universitaria, 04510 México D. F., Mexico; iCenters for Disease Control and Prevention Nigeria, Abuja, Nigeria; jInfectious Diseases Pathology Branch, Division of High-Consequence Pathogens and Pathology, National Center for Emerging Zoonotic Infectious Diseases, Centers for Disease Control and Prevention, Atlanta, GA 30333; kDepartment of Veterinary Medicine, Ahmadu Bello University, Samaru, Zaria, Kaduna State, Nigeria; lCenter for Health Studies, Universidad del Valle de Guatemala, 01015, Guatemala City, Guatemala; mLaboratoire National Vétérinaire, B.P. -

Sero-Monitoring of Horses Demonstrates the Equivac® Hev Hendra Virus Vaccine to Be Highly Effective in Inducing Neu-Tralising A
Article Sero-Monitoring of Horses Demonstrates the Equivac® HeV Hendra Virus Vaccine to Be Highly Effective in Inducing Neutralising Antibody Titres Kim Halpin * , Kerryne Graham and Peter A. Durr Australian Centre for Disease Preparedness (ACDP), Commonwealth Scientific and Industrial Research Organisation (CSIRO), 5 Portarlington Road, East Geelong, VIC 3219, Australia; [email protected] (K.G.); [email protected] (P.A.D.) * Correspondence: [email protected]; Tel.: +61-352275510 Abstract: Hendra virus (HeV) is a high consequence zoonotic pathogen found in Australia. The HeV vaccine was developed for use in horses and provides a One Health solution to the prevention of human disease. By protecting horses from infection, the vaccine indirectly protects humans as well, as horses are the only known source of infection for humans. The sub-unit-based vaccine, containing recombinant HeV soluble G (sG) glycoprotein, was released by Pfizer Animal Health (now Zoetis) for use in Australia at the end of 2012. The purpose of this study was to collate post-vaccination serum neutralising antibody titres as a way of assessing how the vaccine has been performing in the field. Serum neutralization tests (SNTs) were performed on serum samples from vaccinated horses submitted to the laboratory by veterinarians. The SNT results have been analysed, together with age, dates of vaccinations, date of sampling and location. Results from 332 horses formed the data Citation: Halpin, K.; Graham, K.; set. Provided horses received at least three vaccinations (consisting of two doses 3–6 weeks apart, Durr, P.A. Sero-Monitoring of Horses and a third dose six months later), horses had high neutralising titres (median titre for three or more Demonstrates the Equivac® HeV vaccinations was 2048), and none tested negative. -

Analysis of a Reaction Diffusion Model for a Reservoir Supported Spread
Analysis of a Reaction Diffusion Model for a Reservoir Supported Spread of Infectious Disease W.E. Fitzgibbon and J.J. Morgan Department of Mathematics University of Houston Houston, TX 77204, USA Abstract Motivated by recent outbreaks of the Ebola Virus, we are concerned with the role that a vector reservoir plays in supporting the spatio-temporal spread of a highly lethal disease through a host population. In our context, the reservoir is a species capable of harboring and sustaining the pathogen. We develop models that describe the horizontal spread of the disease among the host population when the host population is in contact with the reservoir and when it is not in contact with the host population. These models are of reaction diffusion type, and they are analyzed, and their long term asymptotic behavior is determined. 2000 Mathematics Subject Classification: 34, 35K65, 92B99 Keywords: vector-host, reaction-diffusion, reservoir model, asymptotic behavior. 1 Introduction In what follows we introduce a suite of models of increasing complexity to describe the role of a reservoir in circulating a highly lethal disease among a host population. In our context the reservoir will be a species in which the infectious agent normally lives and multiplies. We assume the reservoir harbors the infectious agent without injury to itself and is capable of transmitting the agent to the host species. We model the outbreak and arXiv:1807.08040v1 [math.AP] 20 Jul 2018 spatio-temporal spread of a highly lethal disease that is initiated and sustained via contact of a host population with an infected reservoir population. -

Operation Outbreak Hamlet’S Story
Operation Outbreak Hamlet’s Story Activity details Age or grade level Activity time This activity is intended for middle and high 45 minutes school teachers to teach public health using the The Junior Disease Detectives, Operation: Handouts Outbreak graphic novel in their classrooms. • Chains of Infection: Story Map Outlines • Story Map Learning objectives At the end of this activity, students should be able to Materials • The Junior Disease Detectives, Operation: • Define zoonotic influenza virus. Outbreak graphic novel • Explain how some influenza viruses in (https://www.cdc.gov/flu/graphicnovel) animals have the potential to cause disease • Internet access in humans, and how some influenza viruses in humans can infect animals. Introduction • Define variant virus and explain how it is a hen an outbreak occurs, disease detectives specific kind of zoonotic influenza virus in W humans; also explain risk factors for variant try to identify what the infectious agent is, how it virus infections in humans. is being transmitted, and who or what is transmitting it by mapping the chain of • Differentiate between direct and indirect infection. The chain of infection provides a transmission and explain the different detailed account of how an infectious agent modes of transmission through which interacts with its host(s) and environment. More influenza viruses can spread between specifically, it shows the sequence of events. It hosts, such as humans and animals. starts when the infectious agent leaves its • Define novel influenza virus and explain reservoir host — the natural habitat (animal or how zoonotic and novel influenza A virus environment). This is where the infectious agent infections on rare occasions can cause normally lives, grows, and multiplies. -

Real Science Review: Zoonotic Diseases
Real Science Review: Zoonotic Diseases Student, Daniel Davidson: Ms. Ribardo, I am so confused about how this COVID-19 epidemic got started. Can you help me understand this? Ms. Ribardo: That’s a really good question, Daniel. I guess the first thing to know is the geographical location where the disease first appeared. In this case, this seems to be in the city of Wuhan, China. The next thing to know is if there is something unusual in that area that might serve as a source of infection. In this case, a wild animal source of the disease was suspected, because this city has many unsanitary meat markets that sell wild animals that are slaughtered at the market. You are going to review a research report that examines the likelihood that this might explain the origin of COVID-19. Does it matter how this disease got started? Think about it. Original Report: Morens, David M. et al. (2020). The origin of COVID-19 and why it matters. Am. J. Trop. Med. Hyg. 103(3), 955-959. Revising author: W. R. Klemm Vocabulary Used in the Original Report Angiotensin-converting enzyme 2: an enzyme in the cell membrane of cells. When certain viruses bind to this enzyme, they break through the membrane to enter and infect the cells. Cell Membrane Receptor: naturally occurring specific molecules, embedded in cell membranes, that can be bound by infectious agents, enzymes, hormones, drugs, or other compounds in ways that alter membrane permeability and cell function. The alterations may be helpful or harmful to cells, depending on the substances being bound. -
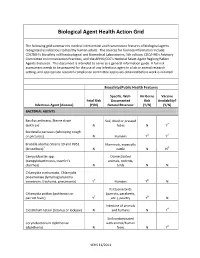
Biological Agent Health Action Grid
Biological Agent Health Action Grid The following grid summarizes medical intervention and transmission features of biological agents recognized as infectious to healthy human adults. The sources for foonote information include: CDC/NIH’s Biosafety in Microbiological and Biomedical Laboratories, 5th edition; CDC/HHS’s Advisory Committee on Immunization Practices; and the APHIS/CDC’s National Select Agent Registry/Select Agents Exclusion. This document is intended to serve as a general information guide. A full risk assessment needs to be prepared for the use of any infectious agent in a lab or animal research setting, and appropriate research compliance committee approvals obtained before work is initiated. Biosafety/Public Health Features Specific, Well- Air-Borne Vaccine Fetal Risk Documented Risk Availability? Infectious Agent (disease) (Y/N) Natural Reservoir (Y/N) (Y/N) BACTERIAL AGENTS Bacillus anthracis, Sterne strain Soil, dried or pressed (anthrax)1 N hides N Y2 Bordetella pertussis (whooping cough or pertussis) N Humans Y3 Y4 Brucella abortus Strains 19 and RB51 Mammals, especially (brucellosis)5 N cattle N N6 Campylobacter spp. Domesticated (campylobacteriosis, traveler's animals, rodents, diarrhea) N birds N N Chlamydia trachomatis, Chlamydia pneumoniae (lymphogranuloma venereum, trachoma, pneumonia) Y7 Humans Y8 N Psittacine birds Chlamydia psittaci (psittacosis or (parrots, parakeets, parrott fever) Y7 etc.), poultry Y8 N Intestine of animals Clostridium tetani (tetanus or lockjaw) N and humans N Y4 Soil contaminated Corynebacterium diphtheriae with animal/human (diphtheria) N feces N Y4 VEHS 11/2011 Specific, Well- Air-Borne Vaccine Fetal Risk Documented Risk Availability? Infectious Agent (disease) (Y/N) Natural Reservoir (Y/N) (Y/N) Francisella tularensis, subspecies Wild animals, esp.