Chemogenetic Profiling Identifies RAD17 As Synthetically Lethal with Checkpoint Kinase Inhibition
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Clinical Candidates Targeting the ATR–CHK1–WEE1 Axis in Cancer
cancers Review Clinical Candidates Targeting the ATR–CHK1–WEE1 Axis in Cancer Lukas Gorecki 1 , Martin Andrs 1,2 and Jan Korabecny 1,* 1 Biomedical Research Center, University Hospital Hradec Kralove, Sokolska 581, 500 05 Hradec Kralove, Czech Republic; [email protected] (L.G.); [email protected] (M.A.) 2 Laboratory of Cancer Cell Biology, Institute of Molecular Genetics of the Czech Academy of Sciences, Videnska 1083, 142 00 Prague, Czech Republic * Correspondence: [email protected]; Tel.: +420-495-833-447 Simple Summary: Selective killing of cancer cells is privileged mainstream in cancer treatment and targeted therapy represents the new tool with a potential to pursue this aim. It can also aid to overcome resistance of conventional chemo- or radio-therapy. Common mutations of cancer cells (defective G1 control) favor inhibiting intra-S and G2/M-checkpoints, which are regulated by ATR–CHK1–WEE1 pathway. The ATR–CHK1–WEE1 axis has produced several clinical candidates currently undergoing clinical trials in phase II. Clinical results from randomized trials by ATR and WEE1 inhibitors warrant ongoing clinical trials in phase III. Abstract: Selective killing of cancer cells while sparing healthy ones is the principle of the perfect cancer treatment and the primary aim of many oncologists, molecular biologists, and medicinal chemists. To achieve this goal, it is crucial to understand the molecular mechanisms that distinguish cancer cells from healthy ones. Accordingly, several clinical candidates that use particular mutations in cell-cycle progressions have been developed to kill cancer cells. As the majority of cancer cells have defects in G1 control, targeting the subsequent intra-S or G2/M checkpoints has also been extensively Citation: Gorecki, L.; Andrs, M.; pursued. -

Wee1 Rather Than Plk1 Is Inhibited by AZD1775 at Therapeutically Relevant Concentrations
cancers Article Wee1 Rather Than Plk1 Is Inhibited by AZD1775 at Therapeutically Relevant Concentrations Angela Flavia Serpico 1,2, Giuseppe D’Alterio 1,2, Cinzia Vetrei 1,2, Rosa Della Monica 1, Luca Nardella 1,2, Roberta Visconti 3 and Domenico Grieco 1,4,* 1 CEINGE Biotecnologie Avanzate, 80145 Naples, Italy; angelafl[email protected] (A.F.S.); [email protected] (G.D.); [email protected] (C.V.); [email protected] (R.D.M.); [email protected] (L.N.) 2 DMMBM, University of Naples “Federico II”, 80131 Naples, Italy 3 IEOS, CNR, 80131 Naples, Italy; [email protected] 4 Department of Pharmacy, University of Naples “Federico II”, 80131 Naples, Italy * Correspondence: [email protected] Received: 24 April 2019; Accepted: 10 June 2019; Published: 13 June 2019 Abstract: Wee1 kinase is an inhibitor of cyclin-dependent kinase (cdk)s, crucial cell cycle progression drivers. By phosphorylating cdk1 at tyrosine 15, Wee1 inhibits activation of cyclin B-cdk1 (Cdk1), preventing cells from entering mitosis with incompletely replicated or damaged DNA. Thus, inhibiting Wee1, alone or in combination with DNA damaging agents, can kill cancer cells by mitotic catastrophe, a tumor suppressive response that follows mitosis onset in the presence of under-replicated or damaged DNA. AZD1775, an orally available Wee1 inhibitor, has entered clinical trials for cancer treatment following this strategy, with promising results. Recently, however, AZD1775 has been shown to inhibit also the polo-like kinase homolog Plk1 in vitro, casting doubts on its mechanism of action. Here we asked whether, in the clinically relevant concentration range, AZD1775 inhibited Wee1 or Plk1 in transformed and non-transformed human cells. -

Application of a MYC Degradation
SCIENCE SIGNALING | RESEARCH ARTICLE CANCER Copyright © 2019 The Authors, some rights reserved; Application of a MYC degradation screen identifies exclusive licensee American Association sensitivity to CDK9 inhibitors in KRAS-mutant for the Advancement of Science. No claim pancreatic cancer to original U.S. Devon R. Blake1, Angelina V. Vaseva2, Richard G. Hodge2, McKenzie P. Kline3, Thomas S. K. Gilbert1,4, Government Works Vikas Tyagi5, Daowei Huang5, Gabrielle C. Whiten5, Jacob E. Larson5, Xiaodong Wang2,5, Kenneth H. Pearce5, Laura E. Herring1,4, Lee M. Graves1,2,4, Stephen V. Frye2,5, Michael J. Emanuele1,2, Adrienne D. Cox1,2,6, Channing J. Der1,2* Stabilization of the MYC oncoprotein by KRAS signaling critically promotes the growth of pancreatic ductal adeno- carcinoma (PDAC). Thus, understanding how MYC protein stability is regulated may lead to effective therapies. Here, we used a previously developed, flow cytometry–based assay that screened a library of >800 protein kinase inhibitors and identified compounds that promoted either the stability or degradation of MYC in a KRAS-mutant PDAC cell line. We validated compounds that stabilized or destabilized MYC and then focused on one compound, Downloaded from UNC10112785, that induced the substantial loss of MYC protein in both two-dimensional (2D) and 3D cell cultures. We determined that this compound is a potent CDK9 inhibitor with a previously uncharacterized scaffold, caused MYC loss through both transcriptional and posttranslational mechanisms, and suppresses PDAC anchorage- dependent and anchorage-independent growth. We discovered that CDK9 enhanced MYC protein stability 62 through a previously unknown, KRAS-independent mechanism involving direct phosphorylation of MYC at Ser . -

Therapeutic Co-Targeting of WEE1 and ATM Downregulates PD-L1 Expression in Pancreatic Cancer
pISSN 1598-2998, eISSN 2005-9256 Cancer Res Treat. 2020;52(1):149-166 https://doi.org/10.4143/crt.2019.183 Original Article Open Access Therapeutic Co-targeting of WEE1 and ATM Downregulates PD-L1 Expression in Pancreatic Cancer Mei Hua Jin, MD, MS1 Purpose Ah-Rong Nam, MS1 Pancreatic cancer (PC) is one of the most lethal cancers worldwide, but there are currently no effective treatments. The DNA damage response (DDR) is under investigation for the Ji Eun Park, MS1 development of novel anti-cancer drugs. Since DNA repair pathway alterations have been MS1 Ju-Hee Bang, found frequently in PC, the purpose of this study was to test the DDR-targeting strategy in 1,2 Yung-Jue Bang, MD, PhD PC using WEE1 and ATM inhibitors. Do-Youn Oh, MD, PhD1,2 Materials and Methods We performed in vitro experiments using a total of ten human PC cell lines to evaluate anti- tumor effect of AZD1775 (WEE1 inhibitor) alone or combination with AZD0156 (ATM inhi- bitor). We established Capan-1–mouse model for in vivo experiments to confirm our findings. Results In our research, we found that WEE1 inhibitor (AZD1775) as single agent showed anti-tumor 1Cancer Research Institute, Seoul National effects in PC cells, however, targeting WEE1 upregulated p-ATM level. Here, we observed University College of Medicine, Seoul, that co-targeting of WEE1 and ATM acted synergistically to reduce cell proliferation and 2Department of Internal Medicine, Seoul migration, and to induce DNA damage in vitro. Notably, inhibition of WEE1 or WEE1/ATM National University Hospital, Seoul, Korea downregulated programmed cell death ligand 1 expression by blocking glycogen synthase kinase-3! serine 9 phosphorylation and decrease of CMTM6 expression. -

A Haploid Genetic Screen Identifies the G1/S Regulatory Machinery As a Determinant of Wee1 Inhibitor Sensitivity
A haploid genetic screen identifies the G1/S regulatory machinery as a determinant of Wee1 inhibitor sensitivity Anne Margriet Heijinka, Vincent A. Blomenb, Xavier Bisteauc, Fabian Degenera, Felipe Yu Matsushitaa, Philipp Kaldisc,d, Floris Foijere, and Marcel A. T. M. van Vugta,1 aDepartment of Medical Oncology, University Medical Center Groningen, University of Groningen, 9723 GZ Groningen, The Netherlands; bDivision of Biochemistry, The Netherlands Cancer Institute, 1066 CX Amsterdam, The Netherlands; cInstitute of Molecular and Cell Biology, Agency for Science, Technology and Research, Proteos#3-09, Singapore 138673, Republic of Singapore; dDepartment of Biochemistry, National University of Singapore, Singapore 117597, Republic of Singapore; and eEuropean Research Institute for the Biology of Ageing, University of Groningen, University Medical Center Groningen, 9713 AV Groningen, The Netherlands Edited by Stephen J. Elledge, Harvard Medical School, Boston, MA, and approved October 21, 2015 (received for review March 17, 2015) The Wee1 cell cycle checkpoint kinase prevents premature mitotic Wee1 kinase at tyrosine (Tyr)-15 to prevent unscheduled Cdk1 entry by inhibiting cyclin-dependent kinases. Chemical inhibitors activity (5, 6). Conversely, timely activation of Cdk1 depends on of Wee1 are currently being tested clinically as targeted anticancer Tyr-15 dephosphorylation by one of the Cdc25 phosphatases drugs. Wee1 inhibition is thought to be preferentially cytotoxic in (7–10). When DNA is damaged, the downstream DNA damage p53-defective cancer cells. However, TP53 mutant cancers do not response (DDR) kinases Chk1 and Chk2 inhibit Cdc25 phos- respond consistently to Wee1 inhibitor treatment, indicating the phatases through direct phosphorylation, which blocks Cdk1 existence of genetic determinants of Wee1 inhibitor sensitivity other activation (11–13). -

CPTC-CDK1-1 (CAB079974) Immunohistochemistry
CPTC-CDK1-1 (CAB079974) Uniprot ID: P06493 Protein name: CDK1_HUMAN Full name: Cyclin-dependent kinase 1 Tissue specificity: Isoform 2 is found in breast cancer tissues. Function: Plays a key role in the control of the eukaryotic cell cycle by modulating the centrosome cycle as well as mitotic onset; promotes G2-M transition, and regulates G1 progress and G1-S transition via association with multiple interphase cyclins. Required in higher cells for entry into S-phase and mitosis. Phosphorylates PARVA/actopaxin, APC, AMPH, APC, BARD1, Bcl-xL/BCL2L1, BRCA2, CALD1, CASP8, CDC7, CDC20, CDC25A, CDC25C, CC2D1A, CENPA, CSNK2 proteins/CKII, FZR1/CDH1, CDK7, CEBPB, CHAMP1, DMD/dystrophin, EEF1 proteins/EF-1, EZH2, KIF11/EG5, EGFR, FANCG, FOS, GFAP, GOLGA2/GM130, GRASP1, UBE2A/hHR6A, HIST1H1 proteins/histone H1, HMGA1, HIVEP3/KRC, LMNA, LMNB, LMNC, LBR, LATS1, MAP1B, MAP4, MARCKS, MCM2, MCM4, MKLP1, MYB, NEFH, NFIC, NPC/nuclear pore complex, PITPNM1/NIR2, NPM1, NCL, NUCKS1, NPM1/numatrin, ORC1, PRKAR2A, EEF1E1/p18, EIF3F/p47, p53/TP53, NONO/p54NRB, PAPOLA, PLEC/plectin, RB1, TPPP, UL40/R2, RAB4A, RAP1GAP, RCC1, RPS6KB1/S6K1, KHDRBS1/SAM68, ESPL1, SKI, BIRC5/survivin, STIP1, TEX14, beta-tubulins, MAPT/TAU, NEDD1, VIM/vimentin, TK1, FOXO1, RUNX1/AML1, SAMHD1, SIRT2 and RUNX2. CDK1/CDC2-cyclin-B controls pronuclear union in interphase fertilized eggs. Essential for early stages of embryonic development. During G2 and early mitosis, CDC25A/B/C-mediated dephosphorylation activates CDK1/cyclin complexes which phosphorylate several substrates that trigger at least centrosome separation, Golgi dynamics, nuclear envelope breakdown and chromosome condensation. Once chromosomes are condensed and aligned at the metaphase plate, CDK1 activity is switched off by WEE1- and PKMYT1-mediated phosphorylation to allow sister chromatid separation, chromosome decondensation, reformation of the nuclear envelope and cytokinesis. -

Functional Genetic Screen Identifies Increased Sensitivity to WEE1 Inhibition in Cells with Defects in Fanconi Anemia and HR Pathways
Published OnlineFirst February 11, 2015; DOI: 10.1158/1535-7163.MCT-14-0845 Small Molecule Therapeutics Molecular Cancer Therapeutics Functional Genetic Screen Identifies Increased Sensitivity to WEE1 Inhibition in Cells with Defects in Fanconi Anemia and HR Pathways Marieke Aarts1, Ilirjana Bajrami1, Maria T. Herrera-Abreu1, Richard Elliott1, Rachel Brough1, Alan Ashworth1, Christopher J. Lord1, and Nicholas C. Turner1,2 Abstract WEE1 kinase regulates CDK1 and CDK2 activity to facilitate increased replication stress and forced mitotic entry of S-phase DNA replication during S-phase and to prevent unscheduled cells. Loss of multiple components of the Fanconi anemia entry into mitosis. WEE1 inhibitors synergize with DNA-dam- (FA) and homologous recombination (HR) pathways, in par- aging agents that arrest cells in S-phase by triggering direct ticular DNA helicases, sensitized to WEE1 inhibition. Silencing mitotic entry without completing DNA synthesis, resulting in of FA/HR genes resulted in excessive replication stress and catastrophic chromosome fragmentation and apoptosis. Here, nucleotide depletion following WEE1 inhibition, which ulti- we investigated how WEE1 inhibition could be best exploited mately led to increased unscheduled mitotic entry. Our results for cancer therapy by performing a functional genetic screen to suggest that cancers with defects in FA and HR pathways may be identify novel determinants of sensitivity to WEE1 inhibition. targeted by WEE1 inhibition, providing a basis for a novel Inhibition of kinases that regulate CDK activity, CHK1 and synthetic lethal strategy for cancers harboring FA/HR defects. MYT1, synergized with WEE1 inhibition through both Mol Cancer Ther; 14(4); 865–76. Ó2015 AACR. Introduction activation (5). -

Targeting the WEE1 Kinase As a Molecular Targeted Therapy for Gastric Cancer
www.impactjournals.com/oncotarget/ Oncotarget, Vol. 7, No. 31 Research Paper Targeting the WEE1 kinase as a molecular targeted therapy for gastric cancer Hye-Young Kim1,2, Yunhee Cho1,3, HyeokGu Kang1,3, Ye-Seal Yim1,3, Seok-Jun Kim1,3, Jaewhan Song2, Kyung-Hee Chun1,3 1Department of Biochemistry & Molecular Biology, Yonsei University College of Medicine, Seodaemun-gu, Seoul 03722, Korea 2Department of Biochemistry, College of Life Science and Biotechnology, Seodaemun-gu, Seoul 03722, Korea 3Brain Korea 21 PlusProject for Medical Science, Yonsei University, Seodaemun-gu, Seoul 03722, Korea Correspondence to: Kyung-Hee Chun, email: [email protected] Keywords: WEE1, AZD1775 (MK-1775), 5-FU, Paclitaxel, gastric cancer Received: September 07, 2015 Accepted: May 28, 2016 Published: June 23, 2016 ABSTRACT Wee1 is a member of the Serine/Threonine protein kinase family and is a key regulator of cell cycle progression. It has been known that WEE1 is highly expressed and has oncogenic functions in various cancers, but it is not yet studied in gastric cancers. In this study, we investigated the oncogenic role and therapeutic potency of targeting WEE1 in gastric cancer. At first, higher expression levels of WEE1 with lower survival probability were determined in stage 4 gastric cancer patients or male patients with accompanied lymph node metastasis. To determine the function of WEE1 in gastric cancer cells, we determined that WEE1 ablation decreased the proliferation, migration, and invasion, while overexpression of WEE1 increased these effects in gastric cancer cells. We also validated the clinical application of WEE1 targeting by a small molecule, AZD1775 (MK-1775), which is a WEE1 specific inhibitor undergoing clinical trials. -

Synergy of WEE1 and Mtor Inhibition in Mutant KRAS-Driven
Published OnlineFirst August 18, 2017; DOI: 10.1158/1078-0432.CCR-17-1098 Cancer Therapy: Preclinical Clinical Cancer Research Synergy of WEE1 and mTOR Inhibition in Mutant KRAS-Driven Lung Cancers Josephine Hai1,2, Shengwu Liu1,2, Lauren Bufe1, Khanh Do1,2, Ting Chen1,3, Xiaoen Wang1, Christine Ng4, Shuai Li1,2, Ming-Sound Tsao4, Geoffrey I. Shapiro1,2, and Kwok-Kin Wong1,2,3 Abstract Purpose: KRAS-activating mutations are the most common Results: We demonstrate that combined inhibition of oncogenic driver in non–small cell lung cancer (NSCLC), but mTOR and WEE1 induced potent synergistic cytotoxic efforts to directly target mutant KRAS have proved a formidable effects selectively in KRAS-mutant NSCLC cell lines, delayed challenge. Therefore, multitargeted therapy may offer a plau- human tumor xenograft growth and caused tumor regres- sible strategy to effectively treat KRAS-driven NSCLCs. Here, we sion in a murine lung adenocarcinoma model. Mechanis- evaluate the efficacy and mechanistic rationale for combining tically, we show that inhibition of mTOR potentiates mTOR and WEE1 inhibition as a potential therapy for lung WEE1 inhibition by abrogating compensatory activation of cancers harboring KRAS mutations. DNA repair, exacerbating DNA damage in KRAS-mutant Experimental Design: We investigated the synergistic effect of NSCLC, and that this effect is due in part to reduction in combining mTOR and WEE1 inhibitors on cell viability, apo- cyclin D1. ptosis, and DNA damage repair response using a panel of Conclusions: These findings demonstrate that compro- human KRAS-mutantandwildtypeNSCLCcelllinesand mised DNA repair underlies the observed potent synergy of patient-derived xenograft cell lines. -
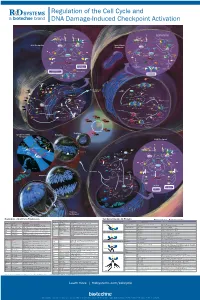
Regulation of the Cell Cycle and DNA Damage-Induced Checkpoint Activation
RnDSy-lu-2945 Regulation of the Cell Cycle and DNA Damage-Induced Checkpoint Activation IR UV IR Stalled Replication Forks/ BRCA1 Rad50 Long Stretches of ss-DNA Rad50 Mre11 BRCA1 Nbs1 Rad9-Rad1-Hus1 Mre11 RPA MDC1 γ-H2AX DNA Pol α/Primase RFC2-5 MDC1 Nbs1 53BP1 MCM2-7 53BP1 γ-H2AX Rad17 Claspin MCM10 Rad9-Rad1-Hus1 TopBP1 CDC45 G1/S Checkpoint Intra-S-Phase RFC2-5 ATM ATR TopBP1 Rad17 ATRIP ATM Checkpoint Claspin Chk2 Chk1 Chk2 Chk1 ATR Rad50 ATRIP Mre11 FANCD2 Ubiquitin MDM2 MDM2 Nbs1 CDC25A Rad50 Mre11 BRCA1 Ub-mediated Phosphatase p53 CDC25A Ubiquitin p53 FANCD2 Phosphatase Degradation Nbs1 p53 p53 CDK2 p21 p21 BRCA1 Ub-mediated SMC1 Degradation Cyclin E/A SMC1 CDK2 Slow S Phase CDC45 Progression p21 DNA Pol α/Primase Slow S Phase p21 Cyclin E Progression Maintenance of Inhibition of New CDC6 CDT1 CDC45 G1/S Arrest Origin Firing ORC MCM2-7 MCM2-7 Recovery of Stalled Replication Forks Inhibition of MCM10 MCM10 Replication Origin Firing DNA Pol α/Primase ORI CDC6 CDT1 MCM2-7 ORC S Phase Delay MCM2-7 MCM10 MCM10 ORI Geminin EGF EGF R GAB-1 CDC6 CDT1 ORC MCM2-7 PI 3-Kinase p70 S6K MCM2-7 S6 Protein Translation Pre-RC (G1) GAB-2 MCM10 GSK-3 TSC1/2 MCM10 ORI PIP2 TOR Promotes Replication CAK EGF Origin Firing Origin PIP3 Activation CDK2 EGF R Akt CDC25A PDK-1 Phosphatase Cyclin E/A SHIP CIP/KIP (p21, p27, p57) (Active) PLCγ PP2A (Active) PTEN CDC45 PIP2 CAK Unwinding RPA CDC7 CDK2 IP3 DAG (Active) Positive DBF4 α Feedback CDC25A DNA Pol /Primase Cyclin E Loop Phosphatase PKC ORC RAS CDK4/6 CDK2 (Active) Cyclin E MCM10 CDC45 RPA IP Receptor -

Torin1-Mediated TOR Kinase Inhibition Reduces Wee1 Levels And
ß 2014. Published by The Company of Biologists Ltd | Journal of Cell Science (2014) 127, 1346–1356 doi:10.1242/jcs.146373 RESEARCH ARTICLE Torin1-mediated TOR kinase inhibition reduces Wee1 levels and advances mitotic commitment in fission yeast and HeLa cells Jane Atkin1, Lenka Halova1, Jennifer Ferguson1, James R. Hitchin2, Agata Lichawska-Cieslar3, Allan M. Jordan2, Jonathon Pines3, Claudia Wellbrock1 and Janni Petersen1,* ABSTRACT target the kinase domain of mTOR, such as Torin1 (Thoreen et al., 2009), mimics the impact of rapamycin treatment in The target of rapamycin (TOR) kinase regulates cell growth and budding yeast, in that they induce autophagy, reduce protein division. Rapamycin only inhibits a subset of TOR activities. Here we synthesis and arrest cell cycle progression in G1 with a reduced show that in contrast to the mild impact of rapamycin on cell division, cell size (Thoreen et al., 2009). These effects of Torin1 blocking the catalytic site of TOR with the Torin1 inhibitor completely established that there are rapamycin-resistant roles for arrests growth without cell death in Schizosaccharomyces pombe.A mTORC1 that are essential for growth and proliferation. Torin1 mutation of the Tor2 glycine residue (G2040D) that lies adjacent to interacts with tryptophan-2239 in the catalytic, active site of the key Torin-interacting tryptophan provides Torin1 resistance, mTOR kinase (Yang et al., 2013). Crucially, this residue is absent confirming the specificity of Torin1 for TOR. Using this mutation, we in other kinases, including the mTOR-related phosphoinositide 3- show that Torin1 advanced mitotic onset before inducing growth kinases (PI3Ks). arrest. In contrast to TOR inhibition with rapamycin, regulation by Here, we describe the isolation of a tor2 mutation that maps to either Wee1 or Cdc25 was sufficient for this Torin1-induced advanced a conserved glycine located next to the key tryptophan (W2239 of mitosis. -

WEE1 Kinase Targeting Combined with DNA-Damaging Cancer Therapy Catalyzes Mitotic Catastrophe
Published OnlineFirst May 11, 2011; DOI: 10.1158/1078-0432.CCR-10-2537 Clinical Cancer Molecular Pathways Research WEE1 Kinase Targeting Combined with DNA-Damaging Cancer Therapy Catalyzes Mitotic Catastrophe Philip C. De Witt Hamer1,3, Shahryar E. Mir2,3, David Noske1,3, Cornelis J.F. Van Noorden4, and Tom Wurdinger€ 1,3,5 Abstract WEE1 kinase is a key molecule in maintaining G2–cell-cycle checkpoint arrest for premitotic DNA repair. Whereas normal cells repair damaged DNA during G1-arrest, cancer cells often have a deficient G1-arrest and largely depend on G2-arrest. The molecular switch for the G2–M transition is held by WEE1 and is pushed forward by CDC25. WEE1 is overexpressed in various cancer types, including glioblastoma and breast cancer. Preclinical studies with cancer cell lines and animal models showed decreased cancer cell viability, reduced tumor burden, and improved survival after WEE1 inhibition by siRNA or small molecule inhibitors, which is enhanced by combination with conventional DNA- damaging therapy, such as radiotherapy and/or cytostatics. Mitotic catastrophe results from premature entry into mitosis with unrepaired lethal DNA damage. As such, cancer cells become sensitized to conventional therapy by WEE1 inhibition, in particular those with insufficient G1-arrest due to deficient p53 signaling, like glioblastoma cells. One WEE1 inhibitor has now reached clinical phase I studies. Dose-limiting toxicity consisted of hematologic events, nausea and/or vomiting, and fatigue. The combination of DNA-damaging cancer therapy with WEE1 inhibition seems to be a rational approach to push cancer cells in mitotic catastrophe. Its safety and efficacy are being evaluated in clinical studies.