Ichthyophis Moustakius Kamei Et Al., 2009
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
Economic and Ecological Implications of Shifting Cultivation in Mizoram, India Environmental Science and Engineering
Environmental Science Vishwambhar Prasad Sati Economic and Ecological Implications of Shifting Cultivation in Mizoram, India Environmental Science and Engineering Environmental Science Series Editors Ulrich Förstner, Technical University of Hamburg-Harburg, Hamburg, Germany Wim H. Rulkens, Department of Environmental Technology, Wageningen, The Netherlands Wim Salomons, Institute for Environmental Studies, University of Amsterdam, Haren, The Netherlands The protection of our environment is one of the most important challenges facing today’s society. At the focus of efforts to solve environmental problems are strategies to determine the actual damage, to manage problems in a viable manner, and to provide technical protection. Similar to the companion subseries Environmental Engineering, Environmental Science reports the newest results of research. The subjects covered include: air pollution; water and soil pollution; renaturation of rivers; lakes and wet areas; biological ecological; and geochemical evaluation of larger regions undergoing rehabilitation; avoidance of environmental damage. The newest research results are presented in concise presentations written in easy to understand language, ready to be put into practice. More information about this subseries at http://www.springer.com/series/3234 Vishwambhar Prasad Sati Economic and Ecological Implications of Shifting Cultivation in Mizoram, India 123 Vishwambhar Prasad Sati Department of Geography and Resource Management Mizoram University (A Central University) Aizawl, Mizoram, India ISSN 1863-5520 ISSN 1863-5539 (electronic) Environmental Science and Engineering ISSN 1431-6250 ISSN 2661-8222 (electronic) Environmental Science ISBN 978-3-030-36601-8 ISBN 978-3-030-36602-5 (eBook) https://doi.org/10.1007/978-3-030-36602-5 © Springer Nature Switzerland AG 2020 This work is subject to copyright. -

ENDOGENOUS RETROVIRUSES in PRIMATES Katherine Brown Bsc
ENDOGENOUS RETROVIRUSES IN PRIMATES Katherine Brown BSc, MSc Thesis submitted to the University of Nottingham for the degree of Doctor of Philosophy July 2015 Abstract Numerous endogenous retroviruses (ERVs) are found in all mammalian genomes, for example, they are the source of approximately 8% of all human and chimpanzee genetic material. These insertions represent retroviruses which have, by chance, integrated into the germline and so are transmitted vertically from parents to offspring. The human genome is rich in ERVs, which have been characterised in some detail. However, in many non-human primates these insertions have not been well- studied. ERVs are subject to the mutation rate of their host, rather than the faster retrovirus mutation rate, so they change much more slowly than exogenous retroviruses. This means ERVs provide a snapshot of the retroviruses a host has been exposed to during its evolutionary history, including retroviruses which are no longer circulating and for which sequence information would otherwise be lost. ERVs have many effects on their hosts; they can be co-opted for functional roles, they provide regions of sequence similarity where mispairing can occur, their insertion can disrupt genes and they provide regulatory elements for existing genes. Accurate annotation and characterisation of these regions is an important step in interpreting the huge amount of genetic information available for increasing numbers of organisms. This project represents an extensive study into the diversity of ERVs in the genomes of primates and related ERVs in rodents. Lagomorphs (rabbits and hares) and tree shrews are also analysed, as the closest relatives of primates and rodents. -

The Mizoram Gazette EXTRA ORDINARY Published Byauthority Regn
The Mizoram Gazette EXTRA ORDINARY Published byAuthority Regn. No. NE-313(MZ) 2006-2008 Rs. 2/- per issue VOL - XXXVII Aizawl, Thursday 11.9.2008 Bhadra 20, S.E. 1930, Issue No. 367 NOTIFICATION No. B. 14016/30/07-LADNC, the 9th September, 2008. LushaiHills District (Administration ofJustice) Rules, 1953, Sec 2 (1) (i) in thuneihna a pek angin Mizoram Governor chuan a hnuaia J... tarian ViUage Council te tan A}·rNEXURE a tarlan ang hian an Boundary a siam a. Tunhma lama heng Village Council te tana boundary 10 siam tawh te chu a thiat nghal ani. It Amaherawh chu heng ramri te hi a tul anga enfiah theih a ni ang. 1 CHA\VILUNGVILLA.GE COUNCIL BOUNDARY 2. TLUNGVEL VILLAGE COUNCIL BOUNDARY 3. DARLAWNG VILLAGE COUNCIL BOUNDARY 4. PHULMAWI VILLAGE COUNCIL BOUNDARY 5. THINGSULTHLIAH, rtllNGSULTLANGNUAM LEHSELINGJOINTVILLAGE COUNCIL BOUNDARY 6. KEPRAN VILLAGE COUNCIL BOUNDARY 7. SAWLENG VILLAGE COUNCIL BOUNDARY 8. N. SERZAWLVILLAGE COUNCIL BOUNDARY 9. SATEEKVILLAGE COUNCIL BOUNDARY 10. MAUBUANG VILLAGE COUNCIL BOUNDARY 11. LENCHIM VILLAGE COUNCIL BOUNDARY 12. RUALLUNG LEH RULCHAWM VILLAGE COUNCIL BOUNDARY 13. DAIDO VILLAGE COUNCIL BOUNDARY 14. N.E.TLANGNUAM VILLAGE COUNCIL BOUNDARY 15. PHUAIBUANG VILLAGE COUNCIL BOUNDARY 16. KHAWLIAN VILLAGE COUNCIl BOUNDARY 17. MUALLUNGTHU VILLAGE COlJNCIL BOUNDARY 18. KELSIH VILLAGE COUNCIL BOUNDARY 19. MELRIATVILLAGE COUNCIL BOUNDARY 20. HUALNGOHMUN VILLAGE COUNCIL BOUNDARY Ex-367/2008 - 2 - 21. SAMTLANG VILLAGE COUNCIL BOUNDARY 22. FALKAWN VILLAGE COUNCIL BOUNDARY 23. N.KHAWLEK VILLAGE COUNCIL BOUNDARY 24. VANBAWNG VILLAGE COUNCIL BOUNDARY 25. LAMHERH VILLAGE COUNCIL BOUNDARY 26. ZAWNGIN VILLAGE COUNCIL BOUNDARY 27. SUANGPUILAWN VILLAGE COUNCIL BOUNDARY 28. SAILAM VILLAGE COUNCIL BOUNDARY 29. -

Hyblaea Puera Cramer (Lepidoptera: Hyblaeidae) Infestation on Tectona Grandis Linn F
© 2020 JETIR November 2020, Volume 7, Issue 11 www.jetir.org (ISSN-2349-5162) Hyblaea puera Cramer (Lepidoptera: Hyblaeidae) infestation on Tectona grandis Linn F. in Aizawl District, Mizoram Lalrinmawia1, Lalnuntluanga2 and Lalramliana3 1,2 Department of Environmental Science, School of Earth Sciences & Natural Resources Management Mizoram University , 3Deparment of Zoology, PUC. Abstract: Seasonal activity of Lepidopteran insect of Hyblaea puera Cramer conducted during three years during of 2016 - 2018 on two aspects of eastern and western sites of Aizawl District, Mizoram. Hyblaea puera Cramer is the most wide spread and serious pest. Outbreaks occur almost every year in India over extensive areas. During these outbreaks in the early flushing period of teak, trees usually suffer a total defoliation, sometimes there is partial defoliation later in the growth season. The present investigation revealed that for bringing out a systematic documentation regarding the damage caused by different insect pests attacking teak plantation in area as well as to find out the relationship of different climatic factors with their incidence. Keywords: Geographical bearing , Hyblaea puera Cramer, Insect Pests, Infestation, Tectona grandis Linn.F. Introduction Teak (Tectona grandis L.f.), a valuable timber species, is attacked by a number of insect pests Mathur,(1960); Mathur and Singh,(1960) ; Baksha,(1990), (1993); Chaiglom,(1975); Menon,(1963). But only one insects-teak defoliator, Hyblaea puera Cramer cause major defoliation of teak in Mizoram. About 187 insects species have been found feeding on living Teak tree in India, Hutacharern and Tubtim, (1995) Amongst the foliage feeders, the teak defoliator, Hyblaea puera Cramer (Hyblaeidae, Lepidoptera) and teak skeletonizer, Eutectona machaeralis Walker (Pyralidae: Lepidoptera) are the most widespread and serious pests. -

Morphotectonic Studies of the Tuirini Drainage Basin:A Remote
International Journal of Geology, Earth & Environmental Sciences ISSN: 2277-2081 (Online) An Open Access, Online International Journal Available at http://www.cibtech.org/jgee.htm 2016 Vol. 6 (1) January-April, pp. 54-65/Ahmed and Rao. Research Article MORPHOTECTONIC STUDIES OF THE TUIRINI DRAINAGE BASIN: A REMOTE SENSING AND GEOGRAPHIC INFORMATION SYSTEM PERSPECTIVE *Fuzal Ahmed and K. Srinivasa Rao Department of Geology, School of Earth Sciences and Natural Resources Management, Mizoram University, Aizawl – 796004, Mizoram, India *Author for Correspondence ABSTRACT Morphotectonic indices have been proven to be useful tools in evaluating the degree and nature of tectonic activity in a specific area, and are commonly used to identify areas affected by recent tectonic deformation. Remote sensing and GIS techniques are considered to be suitable for identifying and quantifying the effects of neotectonic activity over a large area. The analysis has been carried out by using remote sensing and GIS tools in order to understand the ongoing tectonic changes of the terrain in response to neotectonic activities. The study area exhibits complex topography as a result of folding and faulting of sedimentary sequences, and the drainage system is controlled by underlying lithology and geological structures. The basin is elongated in shape due to the combined effects of thrusting and folding in the area. As a whole the terrain is tilted towards the west with asymmetric nature of the basin. The overall results reflect that the Tuirini drainage area is tectonically active in nature. Keywords: Morphotectonic Indices, Neotectonism, GIS, Tuirini River, Mizoram INTRODUCTION The Himalayan mountain belt was evolved during the late Cenozoic deformation caused by convergence collision between the Indian and the Eurasian plates (Molnar and Tapponnier, 1975), which represents one of the youngest and largest foreland belts in the world. -

Western Ghats & Sri Lanka Biodiversity Hotspot
Ecosystem Profile WESTERN GHATS & SRI LANKA BIODIVERSITY HOTSPOT WESTERN GHATS REGION FINAL VERSION MAY 2007 Prepared by: Kamal S. Bawa, Arundhati Das and Jagdish Krishnaswamy (Ashoka Trust for Research in Ecology & the Environment - ATREE) K. Ullas Karanth, N. Samba Kumar and Madhu Rao (Wildlife Conservation Society) in collaboration with: Praveen Bhargav, Wildlife First K.N. Ganeshaiah, University of Agricultural Sciences Srinivas V., Foundation for Ecological Research, Advocacy and Learning incorporating contributions from: Narayani Barve, ATREE Sham Davande, ATREE Balanchandra Hegde, Sahyadri Wildlife and Forest Conservation Trust N.M. Ishwar, Wildlife Institute of India Zafar-ul Islam, Indian Bird Conservation Network Niren Jain, Kudremukh Wildlife Foundation Jayant Kulkarni, Envirosearch S. Lele, Centre for Interdisciplinary Studies in Environment & Development M.D. Madhusudan, Nature Conservation Foundation Nandita Mahadev, University of Agricultural Sciences Kiran M.C., ATREE Prachi Mehta, Envirosearch Divya Mudappa, Nature Conservation Foundation Seema Purshothaman, ATREE Roopali Raghavan, ATREE T. R. Shankar Raman, Nature Conservation Foundation Sharmishta Sarkar, ATREE Mohammed Irfan Ullah, ATREE and with the technical support of: Conservation International-Center for Applied Biodiversity Science Assisted by the following experts and contributors: Rauf Ali Gladwin Joseph Uma Shaanker Rene Borges R. Kannan B. Siddharthan Jake Brunner Ajith Kumar C.S. Silori ii Milind Bunyan M.S.R. Murthy Mewa Singh Ravi Chellam Venkat Narayana H. Sudarshan B.A. Daniel T.S. Nayar R. Sukumar Ranjit Daniels Rohan Pethiyagoda R. Vasudeva Soubadra Devy Narendra Prasad K. Vasudevan P. Dharma Rajan M.K. Prasad Muthu Velautham P.S. Easa Asad Rahmani Arun Venkatraman Madhav Gadgil S.N. Rai Siddharth Yadav T. Ganesh Pratim Roy Santosh George P.S. -

The Mizoram Gazette Published by Author,Ity
Regd. No. NE 907 The Mizoram Gazette Published by Author,ity Vol XUI Aizawl Friday 2.11.1984 Kartika 11 S.B. 1906 Issue No. 44 , ;t)vernment of ,Mizoram PA!:tT I ,t.,;�"P8,;-,�,nents, Postings, Transfers, Powers, Leave and other F\;rsonal Notices and OrGers. ORDERS BY THE LT. GOV ERNOR (ADMINISTRATOR) NOTIFICATIONS , No.B. 20022j 1/84-EDN/3, the 29th October, 1984. The Lt. Governor of Mizoram is pleased to constitute a Co-ordination Committee of Archives with the following members and under the terms of reference shown below with immediate effect and until further order:- 1. 1) Chief Secretary Govt. of Mizoram. Chairman ...- 2) Deputy Commissioner Aizawl. Member 3) Education Secretary, Govt. of Mizoram. -do- Director of Education, Mizoram. -do- 54)) Dy Director Education. i/c Adult & Culture, Mizoram, Aizawl. -do- 6) President, District Council Court. -do- -7) Director of Agriculture, Mizoram. -do- '\. 8) Chief En,2ineer; P.W.D. Aizawl. - -do- 8) Su b-Diyisional Education Officer, Aizawl West.- ' -do- lO) Senior Executive Secretary, Mizoram Presbyterian Church. -do- l l) Representative of the Mizoram Baptist MIssion Church Lunglei -do-- 12) Representative of the Mizoram Roman Catholic Church, .Aizawl. --do- 13� Renresentative of the Mizoram Salvation Army, Aizawl. -do- 14) Superintendent of Archives -do- 15) Curator. J\1izoram· State Museum Mem ber Secretary. R-44/84 2 ' II. The function of the Co-ordi nation Committee of Archives is to implement the (i)main fWlctions of a Shte Record Office as follows:- To conccntrak in a single repos itory a]l the non current records, both , (a) Confidential and non-confidential of the State �ecretariat and the s ubor dingte t:1uthorities to it whether at the headqua'rters or elsewhere inc1udin£r District Divis'on' and coUectorate records and similar records I of the igh Court and othtr cou t; H r (b) To house th:m in a properly equipped building: (c) To arr.]I1.;e (IIld clarify them on Scientific principles; (d) To take such measures, as are required; for their preservation and reha- bilitation; . -

Biogeographic Analysis Reveals Ancient Continental Vicariance and Recent Oceanic Dispersal in Amphibians ∗ R
Syst. Biol. 63(5):779–797, 2014 © The Author(s) 2014. Published by Oxford University Press, on behalf of the Society of Systematic Biologists. All rights reserved. For Permissions, please email: [email protected] DOI:10.1093/sysbio/syu042 Advance Access publication June 19, 2014 Biogeographic Analysis Reveals Ancient Continental Vicariance and Recent Oceanic Dispersal in Amphibians ∗ R. ALEXANDER PYRON Department of Biological Sciences, The George Washington University, 2023 G Street NW, Washington, DC 20052, USA; ∗ Correspondence to be sent to: Department of Biological Sciences, The George Washington University, 2023 G Street NW, Washington, DC 20052, USA; E-mail: [email protected]. Received 13 February 2014; reviews returned 17 April 2014; accepted 13 June 2014 Downloaded from Associate Editor: Adrian Paterson Abstract.—Amphibia comprises over 7000 extant species distributed in almost every ecosystem on every continent except Antarctica. Most species also show high specificity for particular habitats, biomes, or climatic niches, seemingly rendering long-distance dispersal unlikely. Indeed, many lineages still seem to show the signature of their Pangaean origin, approximately 300 Ma later. To date, no study has attempted a large-scale historical-biogeographic analysis of the group to understand the distribution of extant lineages. Here, I use an updated chronogram containing 3309 species (~45% of http://sysbio.oxfordjournals.org/ extant diversity) to reconstruct their movement between 12 global ecoregions. I find that Pangaean origin and subsequent Laurasian and Gondwanan fragmentation explain a large proportion of patterns in the distribution of extant species. However, dispersal during the Cenozoic, likely across land bridges or short distances across oceans, has also exerted a strong influence. -
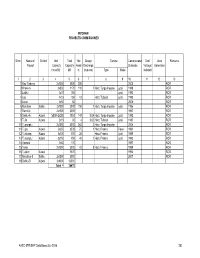
AHEC-IITR/SHP Data Base/July 2016 202 MIZORAM IDENTIFIED FUTURE PROJECTS
MIZORAM PROJECTS COMMISSIONED Sl no. Name of District Unit Total Net Design Turbine Commissionin Grid/ Units Remarks Project Capacity Capacity Head Discharge Schedule Voltage/ Generated (no.xkW) kW m (cumecs) Type Make Isolated 12 3 4 567 8 910111213 1 Kau-Tlabung 2x1500 3000 200 2005 ROR 2 Khawiva 3x350 1170 110 1 Horz. Turgo Impulse Jyoti 1988 ROR 3 Laililui 1x15 150 Jyoti 1993 ROR 4 Lao 1x15 150 13 Horz. Tubular Jyoti 1992 ROR 5 Leiva 1x50 50 2000 ROR 6 Maicham Saiha 2x1000 2000 150 1 Horz. Turgo Impulse Jyoti 1996 ROR 7 Ramrilui 2x1500 3000 1997 ROR 8 Serilui-A Aizwal 1x500+2x250 1000 148 0.04 Horz. Turgo Impulse Jyoti 1992 ROR 9 Tuila Aizwal 2x15 30 4 0.02 Horz. Tubular Jyoti 1987 ROR 10 Tuipanglui 2x1500 3000 243 2 Horz. Turgo Impulse 2004 ROR 11 Tuipui Aizwal 2x250 2500 75 1 Horz. Francis Flovel 1991 ROR 12 Tuirivang Aizwal 3x100 300 28 Horz. Francis Jyoti 1989 ROR 13 Tuisumpui Aizwal 3x150 450 40 1 Horz. Francis Jyoti 1992 ROR 14 Vavralui 1x50 170 1997 ROR 15 Teirei 2x1500 3000 42 8 Horz. Francis 1999 ROR 16 Tuidum Aizwal 1500 1992 ROR 17 Maicham-II Saiha 2x1500 3000 2007 ROR 18 Serilui-B Aizwal 3x4000 12000 Total = 36470 AHEC-IITR/SHP Data Base/July 2016 202 MIZORAM IDENTIFIED FUTURE PROJECTS Sl no. Name of Name of Name of Category Name of Capacity Head Discharge Remarks Verified Project State District of Proj river/ in in in * canal kW m m3/sec 1 Ainaklui Mizoram Chhimtuipui ROR 50 40 Under Investigation 2 Bualanukham Mizoram Chhimtuipui ROR 50 22 Under Investigation 3 Chawngte Mizoram ROR 150 30 Under Investigation 4 Cheului Mizoram Chhimtuipui ROR 50 40 Under Investigation 5 Chiahpui Mizoram Aizawl ROR 20 20 Under Investigation 6 Chikurlui Mizoram Chhimtuipui ROR 15 20 Under Investigation 7 Chunghkum Mizoram ROR 100 100 Under Investigation 8 Dhaleswari -I Mizoram DB Dhaleswari Brk. -

Amphibia: Gymnophiona)
March 1989] HERPETOLOGICA 23 viridiflavus (Dumeril et Bibron, 1841) (Anura Hy- SELANDER, R. K., M. H. SMITH, S. Y. YANG, W. E. peroliidae) en Afrique centrale. Monit. Zool. Itali- JOHNSON, AND J. B. GENTRY. 1971. Biochemical ano, N.S., Supple. 1:1-93. polymorphism and systematics in the genus Pero- LYNCH, J. D. 1966. Multiple morphotypy and par- myscus. I. Variation in the old-field mouse (Pero- allel polymorphism in some neotropical frogs. Syst. myscus polionotus). Stud. Genetics IV, Univ. Texas Zool. 15:18-23. Publ. 7103:49-90. MAYR, E. 1963. Animal Species and Evolution. Har- SICILIANO, M. J., AND C. R. SHAW. 1976. Separation vard University Press, Cambridge. and localization of enzymes on gels. Pp. 184-209. NEI, M. 1972. Genetic distance between popula- In I. Smith (Ed.), Chromatographic and Electro- tions. Am. Nat. 106:283-292. phoretic Techniques, Vol. 2, 4th ed. Williams Hei- RESNICK, L. E., AND D. L. JAMESON. 1963. Color nemann' Medical Books, London. polymorphism in Pacific treefrogs. Science 142: ZIMMERMAN, H., AND E. ZIMMERMAN. 1987. Min- 1081-1083. destanforderungen fur eine artgerechte Haltung SAVAGE, J. M., AND S. B. EMERSON. 1970. Central einiger tropischer Anurenarten. Zeit. Kolner Zoo American frogs allied to Eleutherodactylusbrans- 30:61-71. fordii (Cope): A problem of polymorphism. Copeia 1970:623-644. Accepted: 12 March 1988 Associate Editor: John Iverson SCHIOTZ, A. 1971. The superspecies Hyperoliusvir- idiflavus (Anura). Vidensk. Medd. Dansk Natur- hist. Foren. 134:21-76. Herpetologica,45(1), 1989, 23-36 ? 1989 by The Herpetologists'League, Inc. ON THE STATUS OF NECTOCAECILIA FASCIATA TAYLOR, WITH A DISCUSSION OF THE PHYLOGENY OF THE TYPHLONECTIDAE (AMPHIBIA: GYMNOPHIONA) MARK WILKINSON Museum of Zoology and Department of Biology, University of Michigan, Ann Arbor, MI 48109, USA ABSTRACT: Nectocaecilia fasciata Taylor is a junior synonym of Chthonerpetonindistinctum Reinhardt and Liitken. -

Sertoli Cells in the Testis of Caecilians, Ichthyophis Tricolor and Uraeotyphlus Cf. Narayani (Amphibia: Gymnophiona): Light and Electron Microscopic Perspective
JOURNAL OF MORPHOLOGY 258:317–326 (2003) Sertoli Cells in the Testis of Caecilians, Ichthyophis tricolor and Uraeotyphlus cf. narayani (Amphibia: Gymnophiona): Light and Electron Microscopic Perspective Mathew Smita,1 Oommen V. Oommen,1* Jancy M. George,2 and M.A. Akbarsha2 1Department of Zoology, University of Kerala, Kariavattom, Thiruvananthapuram 695 581, Kerala, India 2Departmnt of Animal Sciences, Bharathidasan University, Thiruchirappalli 620 024, Tamilnadu, India ABSTRACT The caecilians have evolved a unique pat- that surround the cyst/follicle (Fraile et al., 1990; tern of cystic spermatogenesis in which cysts representing Saez et al., 1990; Grier, 1993; Koulish et al., 2002). different stages in spermatogenesis coexist in a testis lob- The follicle cells are known as Sertoli cells when ule. We examined unsettled issues relating to the organi- spermatids form a bundle. Their apical tips become zation of the caecilian testis lobules, including the occur- embedded in the crypts formed by invaginations of rence of a fatty matrix, the possibility of both peripheral and central Sertoli cells, the origin of Sertoli cells from the follicle cell membrane and point towards its follicular cells, and the disengagement of older Sertoli nucleus. This arrangement resembles the Sertoli cells to become loose central Sertoli cells. We subjected the cell / germ cell association in amniotes (Lofts, 1974; testis of Ichthyophis tricolor (Ichthyophiidae) and Uraeo- Bergmann et al., 1982). In anamniotes, with the typhlus cf. narayani (Uraeotyphliidae) from the Western progression of spermatogenesis, isogenic spermato- Ghats of Kerala, India, to light and transmission electron zoa are produced and released by rupture of the cyst microscopic studies. -

Phallus Morphology in Caecilians (Amphibia, Gymnophiona) and Its Systematic Utility
Bull. nat. Hist. Mus. Lond. (Zool.) 68(2): 143–154 Issued 28 November 2002 Phallus morphology in caecilians (Amphibia, Gymnophiona) and its systematic utility DAVID J. GOWER AND MARK WILKINSON Department of Zoology, The Natural History Museum, London SW7 5BD, UK. email addresses: [email protected], [email protected] CONTENTS Introduction ............................................................................................................................................................................. 143 Abbreviation used in text ..................................................................................................................................................... 144 Abbreviations used in figures .............................................................................................................................................. 144 Morphology ............................................................................................................................................................................. 144 Disposition of the cloaca ..................................................................................................................................................... 144 Divisions of the cloaca ........................................................................................................................................................ 146 Urodeum .............................................................................................................................................................................