Genetic Stock Structure of the Swordfish (Xiphias Gladius) Inferred
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Atlantic Cod (Gadus Morhua) Off Newfoundland and Labrador Determined from Genetic Variation
COSEWIC Assessment and Update Status Report on the Atlantic Cod Gadus morhua Newfoundland and Labrador population Laurentian North population Maritimes population Arctic population in Canada Newfoundland and Labrador population - Endangered Laurentian North population - Threatened Maritimes population - Special Concern Arctic population - Special Concern 2003 COSEWIC COSEPAC COMMITTEE ON THE STATUS OF COMITÉ SUR LA SITUATION ENDANGERED WILDLIFE DES ESPÈCES EN PÉRIL IN CANADA AU CANADA COSEWIC status reports are working documents used in assigning the status of wildlife species suspected of being at risk. This report may be cited as follows: COSEWIC 2003. COSEWIC assessment and update status report on the Atlantic cod Gadus morhua in Canada. Committee on the Status of Endangered Wildlife in Canada. Ottawa. xi + 76 pp. Production note: COSEWIC would like to acknowledge Jeffrey A. Hutchings for writing the update status report on the Atlantic cod Gadus morhua, prepared under contract with Environment Canada. For additional copies contact: COSEWIC Secretariat c/o Canadian Wildlife Service Environment Canada Ottawa, ON K1A 0H3 Tel.: (819) 997-4991 / (819) 953-3215 Fax: (819) 994-3684 E-mail: COSEWIC/[email protected] http://www.cosewic.gc.ca Également disponible en français sous le titre Rapport du COSEPAC sur la situation de la morue franche (Gadus morhua) au Canada Cover illustration: Atlantic Cod — Line drawing of Atlantic cod Gadus morhua by H.L. Todd. Image reproduced with permission from the Smithsonian Institution, NMNH, Division of Fishes. Her Majesty the Queen in Right of Canada, 2003 Catalogue No.CW69-14/311-2003-IN ISBN 0-662-34309-3 Recycled paper COSEWIC Assessment Summary Assessment summary — May 2003 Common name Atlantic cod (Newfoundland and Labrador population) Scientific name Gadus morhua Status Endangered Reason for designation Cod in the inshore and offshore waters of Labrador and northeastern Newfoundland, including Grand Bank, having declined 97% since the early 1970s and more than 99% since the early 1960s, are now at historically low levels. -

Status of Shortnose Sturgeon in the Potomac River
Final Report Status of Shortnose Sturgeon in the Potomac River PART I – FIELD STUDIES Report prepared by: Boyd Kynard (Principal Investigator) Matthew Breece and Megan Atcheson (Project Leaders) Micah Kieffer (Co-Investigator) U. S. Geological Survey, Biological Resources Division Leetown Science Center S. O. Conte Anadromous Fish Research Center Turners Falls, Massachusetts 01376 and Mike Mangold (Assistant Project Leader) U. S. Fish and Wildlife Service, Maryland Fishery Resources Office 177 Admiral Cochrane Drive Annapolis, Maryland 21401 Report prepared for: National Park Service National Capital Region Washington, D.C USGS Natural Resources Preservation Project E 2002-7 NPS Project Coordinator – Jim Sherald BRD Project Coordinator – Ed Pendelton July 20, 2007 Gravid shortnose sturgeon female captured at river kilometer 63 on the Potomac River. Project leader, Matthew Breece (USGS), is shown with the fish on March 23, 2006. Summary Field studies during more than 3 years (March 2004–July 2007) collected data on life history of Potomac River shortnose sturgeon Acipenser brevirostrum to understand their biological status in the river. We sampled intensively for adults using gill nets, but captured only one adult in 2005. Another adult was captured in 2006 by a commercial fisher. Both fish were females with excellent body and fin condition, both had mature eggs, and both were telemetry- tagged to track their movements. The lack of capturing adults, even when intensive netting was guided by movements of tracked fish, indicated abundance of the species was less than in any river known with a sustaining population of the species. Telemetry tracking of the two females (one during September 2005–July 2007, one during March 2006–February 2007) found they remained in the river for all the year, not for just a few months like sturgeons on a coastal migration. -

Little Fish, Big Impact: Managing a Crucial Link in Ocean Food Webs
little fish BIG IMPACT Managing a crucial link in ocean food webs A report from the Lenfest Forage Fish Task Force The Lenfest Ocean Program invests in scientific research on the environmental, economic, and social impacts of fishing, fisheries management, and aquaculture. Supported research projects result in peer-reviewed publications in leading scientific journals. The Program works with the scientists to ensure that research results are delivered effectively to decision makers and the public, who can take action based on the findings. The program was established in 2004 by the Lenfest Foundation and is managed by the Pew Charitable Trusts (www.lenfestocean.org, Twitter handle: @LenfestOcean). The Institute for Ocean Conservation Science (IOCS) is part of the Stony Brook University School of Marine and Atmospheric Sciences. It is dedicated to advancing ocean conservation through science. IOCS conducts world-class scientific research that increases knowledge about critical threats to oceans and their inhabitants, provides the foundation for smarter ocean policy, and establishes new frameworks for improved ocean conservation. Suggested citation: Pikitch, E., Boersma, P.D., Boyd, I.L., Conover, D.O., Cury, P., Essington, T., Heppell, S.S., Houde, E.D., Mangel, M., Pauly, D., Plagányi, É., Sainsbury, K., and Steneck, R.S. 2012. Little Fish, Big Impact: Managing a Crucial Link in Ocean Food Webs. Lenfest Ocean Program. Washington, DC. 108 pp. Cover photo illustration: shoal of forage fish (center), surrounded by (clockwise from top), humpback whale, Cape gannet, Steller sea lions, Atlantic puffins, sardines and black-legged kittiwake. Credits Cover (center) and title page: © Jason Pickering/SeaPics.com Banner, pages ii–1: © Brandon Cole Design: Janin/Cliff Design Inc. -

Reproductive Biology of American Shad, Alosa Sapidissima, in the Mattaponi River
W&M ScholarWorks Dissertations, Theses, and Masters Projects Theses, Dissertations, & Master Projects 2004 Reproductive Biology of American Shad, Alosa sapidissima, in the Mattaponi River Aaron Reid Hyle College of William and Mary - Virginia Institute of Marine Science Follow this and additional works at: https://scholarworks.wm.edu/etd Part of the Fresh Water Studies Commons, Oceanography Commons, and the Zoology Commons Recommended Citation Hyle, Aaron Reid, "Reproductive Biology of American Shad, Alosa sapidissima, in the Mattaponi River" (2004). Dissertations, Theses, and Masters Projects. Paper 1539617824. https://dx.doi.org/doi:10.25773/v5-nryc-fp96 This Thesis is brought to you for free and open access by the Theses, Dissertations, & Master Projects at W&M ScholarWorks. It has been accepted for inclusion in Dissertations, Theses, and Masters Projects by an authorized administrator of W&M ScholarWorks. For more information, please contact [email protected]. REPRODUCTIVE BIOLOGY OF AMERICAN SHAD, ALOSA SAPIDISSIMA, IN THE MATTAPONI RIVER A Thesis Presented to The Faculty of the School of Marine Science The College of William and Mary in Virginia In Partial Fulfillment Of the Requirements for the Degree of Master of Science by Aaron Reid Hyle 2004 APPROVAL SHEET This thesis is submitted in partial fulfillment of The requirements for the degree of Master of Science A a r o i S j ^ Approved, January 2004 John EXOlney, Ph.D. Robert J. Latour, Ph.D. Herbert M. Austin, Ph.D. TABLE OF CONTENTS TABLE OF CONTENTS...................................................................... -

Report to The
Final Report to the NOAA Fisheries - Northeast Region Cooperative Research Partners Initiative on the Maine - New Hampshire Inshore Groundfish Trawl Survey July 2001 – June 2002 Final Report Fall 2001 and Spring 2002 Maine – New Hampshire Inshore Trawl Survey Submitted to the NOAA Fisheries-Northeast Region, Cooperative Research Partners Initiative (Contract 50-EANF-1-00013) By Sally A. Sherman, Vincent Manfredi, Jeanne Brown, Hannah Smith, and John Sowles Maine Department of Marine Resources Douglas E. Grout New Hampshire Fish and Game Department Donald W. Perkins, Jr. Gulf of Maine Aquarium Development Corporation And Robert Tetrault T/R Fish Inc. F/V Tara Lynn and F/V Robert Michael March 2003 Technical Research Document 03/1 TABLE OF CONTENTS Acknowledgements iii Executive Summary iv Introduction 1 Materials and Methods 3 Results 10 Fall 2001 Summary 10 Spring 2002 Summary 11 July 2001 Special Survey Summary 12 August 2001 Special Survey Summary 15 Ichthyoplankton 16 Selected Species 17 Atlantic cod (Gadus morhua) 17 Haddock (Melanogrammus aeglefinus) 20 American plaice (Hippoglossoides platessoides) 23 Yellowtail flounder (Limanda ferruginea) 25 Winter flounder (Pseudopluronectes americanus) 27 Goosefish (Lophius americanus) 30 Witch flounder (Glyptocephalus cynoglossus) 32 Atlantic herring (Clupea harengus) 34 Sea scallop (Placopecten magellanicus) 35 American lobster (Homarus americanus) 36 Discussion 39 Literature Cited 42 Appendix A: Individual Station Descriptions A-1 Appendix B: Taxa List B-1 Appendix C: Survey Catch Summaries C-1 Appendix D: Policy on Release of Trawl Survey Data D-1 ii ACKNOWLEDGEMENTS Logistically, this was a complex project that benefited from the assistance of many people. Without their help, the survey could not have been completed. -
Epa Contract No
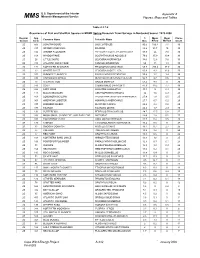
U.S. Department of the Interior Appendix A Minerals Management Service MM S Figures, Maps and Tables Table 4.2.7-6 Occurrence of Fish and Shellfish Species in MDMF Spring Research Trawl Surveys in Nantucket Sound: 1978-2008 Region/ Spp % Mean Mean Cruise Common Name Scientific Name Season Code Occ. #/Tow Wt/Tow Count 2S 503 LONGFIN SQUID LOLIGO PEALEII 90.5 100.4 7.1 30 2S 317 SPIDER CRAB UNCL MAJIDAE 88.2 93.7 10 30 2S 106 WINTER FLOUNDER PSEUDOPLEURONECTES AMERICANUS 87.9 26 7.8 30 2S 108 WINDOWPANE SCOPHTHALMUS AQUOSUS 79.6 35.9 10.4 30 2S 26 LITTLE SKATE LEUCORAJA ERINACEA 78.6 12.6 7.6 30 2S 313 ATLANTIC ROCK CRAB CANCER IRRORATUS 69 13 1.3 30 2S 171 NORTHERN SEAROBIN PRIONOTUS CAROLINUS 68.8 205.4 37.9 30 2S 23 WINTER SKATE LEUCORAJA OCELLATA 60.9 9.3 12.6 30 2S 103 SUMMER FLOUNDER PARALICHTHYS DENTATUS 55.4 1.7 1.4 30 2S 336 CHANNELED WHELK BUSYCOTYPUS CANALICULATUS 54.7 2.7 0.6 30 2S 73 ATLANTIC COD GADUS MORHUA 53.4 9.9 0 30 2S 143 SCUP STENOTOMUS CHRYSOPS 47.9 39.2 7.7 30 2S 322 LADY CRAB OVALIPES OCELLATUS 44.3 5 0.4 30 2S 141 BLACK SEA BASS CENTROPRISTIS STRIATA 30 1.6 0.9 29 2S 163 LONGHORN SCULPIN MYOXOCEPHALUS OCTODECEMSPINOSUS 27.9 1.8 0.5 25 2S 301 AMERICAN LOBSTER HOMARUS AMERICANUS 27.7 0.7 0.2 29 2S 337 KNOBBED WHELK BUSYCON CARICA 26.4 2.4 0.8 29 2S 177 TAUTOG TAUTOGA ONITIS 26.2 1.7 2.9 30 2S 131 BUTTERFISH PEPRILUS TRIACANTHUS 24.7 17.5 0.8 27 2S 338 MOON SNAIL, SHARK EYE, AND BABY-EAR NATICIDAE 24.4 1.8 0.1 27 2S 318 HORSESHOE CRAB LIMULUS POLYPHEMUS 20.7 0.4 0.5 28 2S 176 CUNNER TAUTOGOLABRUS ADSPERSUS 16.4 0.5 -

Feeding Behavior of American Shad During Spawning Migration in the Yo R K River, Virginia
American Fisheries Society Symposium 00:000–000, 2003 © 2003 by the American Fisheries Society Feeding Behavior of American Shad during Spawning Migration in the Yo r k River, Virginia JOHN F. W ALTER, III1 AND JOHN E. OLNEY2 Virginia Institute of Marine Science, School of Marine Science, The College of William and Mary, Gloucester Point, Virginia 23062, USA Abstract.—Throughout its native range, American shad Alosa sapidissima is thought to cease feeding during anadromous migration. We examined the feeding habits of American shad during spawning migration by documenting changes in diet composition and feeding that occur in mature fish in the York River, Virginia. American shad were collected prior to and after spawning at various locations on a gradient from open ocean waters to the freshwater spawning grounds. During the estuarine phase of migration, American shad fed predominantly upon mysid shrimp Neomysis americana and calanoid copepods. Feeding occurred during prespawning migration but decreased on the spawning grounds where plant matter comprised the majority of the stomach contents. After spawning, feeding intensity increased significantly as American shad resumed feeding on mysid shrimp and copepods during downstream migration through estuarine waters. Comparisons of diet composition and feeding intensity between the oceanic environment and the estuary indicate that calanoid copepods and planktonic shrimp dominated the diet in both environments, although feeding intensity was reduced within the estuary during the spawning migration. These results indicate that American shad in the York River feed during anadromous migration, most heavily in the middle estuary where there is an abundance of mysid shrimp. While shad lose somatic weight during migration, indicating insufficient consumption to maintain body condition, the ability to feed during some portion of migration and then soon after spawning may be important in decreasing postspawning mortality. -

Opportunistic Acoustic Telemetry Platforms
NOAA Technical Memorandum NMFS-NE-265 Opportunistic Acoustic Telemetry Platforms: An Update on the Northeast Fisheries Science Center’s Collaborative Monitoring Program in the Gulf of Maine, 2005-2018 by Graham S. Goulette1, James P. Hawkes1, John F. Kocik1, James P. Manning2, Eric Matzen2, Sofie Van Parijs2, Neil Pettigrew3, John Wallinga3, Gayle B. Zydlewski3, Catlin Ames3 US DEPARTMENT OF COMMERCE National Oceanic and Atmospheric Administration National Marine Fisheries Service Northeast Fisheries Science Center Woods Hole, Massachusetts February 2021 NOAA Technical Memorandum NMFS-NE-265 This series represents a secondary level of scientific publishing. All issues employ thorough internal scientific review; some issues employ external scientific review. Reviews are transparent collegial reviews, not anonymous peer reviews. All issues may be cited in formal scientific communications. Opportunistic Acoustic Telemetry Platforms: An Update on the Northeast Fisheries Science Center’s Collaborative Monitoring Program in the Gulf of Maine, 2005-2018 by Graham S. Goulette1, James P. Hawkes1, John F. Kocik1, James P. Manning2, Eric Matzen2, Sofie Van Parijs2, Neil Pettigrew3, John Wallinga3, Gayle B. Zydlewski3, Catlin Ames3 1 NOAA Fisheries Service, Northeast Fisheries Science Center, Maine Field Station, 17 Godfrey Drive - Suite 1, Orono, Maine 04473 2 NOAA Fisheries Service, Northeast Fisheries Science Center, 166 Water Street, Woods Hole, Massachusetts 02543 3 University of Maine, School of Marine Sciences, Orono, Maine 04473 US DEPARTMENT OF COMMERCE Wynn Coggins, ActingSecretary National Oceanic and Atmospheric Administration Benjamin Friedman, Acting Under Secretary National Marine Fisheries Service Paul Doremus, Acting Administrator for Fisheries Northeast Fisheries Science Center Woods Hole, Massachusetts February 2021 Editorial Notes Information Quality Act Compliance: In accordance with section 515 of Public Law 106-554, the Northeast Fisheries Science Center completed both technical and policy reviews for this report. -

Estimating Year-Class Strength in Capelin
NOT TO BE CITED WITHOUT PRIOR REFERENCE TO THE AUTHOR(S) Northwest Atian^ iG Fisheries organization Serial No. N710 NAFO SCR Doc. 83/VI/52 SCIENTIFIC PCOUNCIL MEETING - JUNE 1983 Estimating Year-Class Strength in ^Capelin (Mallotus villosus) From Abiotic Variables by W.C. Leggett l , K. Frank2 and J.E. Carscadden3 Department of Biology, McGill University!, 1205 Avenue Docteur Penfield, Montreal, Quebec H3A 1B1 Marine Ecology Laboratory, Bedford Inst 11 tute of Oceanography, Dartmouth, N.S. B2Y 4A2 3. Department of Fisheries and Oceans, Nor hwest Atlantic Fisheries Centre, P.O. Box 5667, St. Johns, Newfoundland, Canada Al C 5X1 INTRODUCTION . Forecasting recruitment. to mature or exploitable stocks has tong been a central problem in fishery research. Stock-recruitment model< have dominated this field since Ri.ckers (1954) .classic treatment of the subject. These models are l!I based exclusively on biological interactions and fail to incorporate potentially important abiotic influences on recruitment processes. Thi.s may, .in . part, explain their relatively poor predictive p^ lwer, although Walters and Ludwig (1981) have recently demonstrated that the approach is also fra.Gght with statistical difficulties, the principal problem being unknown error in the independent varable (parent stock size) . Several investigators have attempted to account for the unexplained variance in stock- recruitment models by examining the relati lnships between residuals and a variety of abiotic factors. For example, Nelson et al (1979) demonstr ted a significant positive effect of onshore Ekmann transport of larvae on recruitment success in Atlantic menhaden; Bailey (1981) reported a negative relationship between the intensity of offshpre Ekmann transport and year-class-strength in Pacific hake; Leggett (1977) showed that water temperature during the immediate post spawning period was an important cause of residual variance in the stock-recruitment model for American shad (Alosa sapid i ssima) in the Connecticut River; and ,hadwick et al. -

Fish Stocks United Nations Food and Agriculture Organization (FAO)
General situation of world fish stocks United Nations Food and Agriculture Organization (FAO) Contents: 1. Definitions 2. Snapshot of the global situation 3. Short list of "depleted" fish stocks 4. Global list of fish stocks ranked as either "overexploited," "depleted," or recovering by region 1. Definitions Underexploited Undeveloped or new fishery. Believed to have a significant potential for expansion in total production; Moderately exploited Exploited with a low level of fishing effort. Believed to have some limited potential for expansion in total production; Fully exploited The fishery is operating at or close to an optimal yield level, with no expected room for further expansion; Overexploited The fishery is being exploited at above a level which is believed to be sustainable in the long term, with no potential room for further expansion and a higher risk of stock depletion/collapse; Depleted Catches are well below historical levels, irrespective of the amount of fishing effort exerted; Recovering Catches are again increasing after having been depleted 2. Snapshot of the global situation Of the 600 marine fish stocks monitored by FAO: 3% are underexploited 20% are moderately exploited 52% are fully exploited 17% are overexploited 7% are depleted 1% are recovering from depletion Map of world fishing statistical areas monitored by FAO Source: FAO's report "Review of the State of World Marine Fisheries Resources", tables D1-D17, ftp://ftp.fao.org/docrep/fao/007/y5852e/Y5852E23.pdf 3. Fish stocks identified by FAO as falling into its -

Short-Term Mortality in Post-Emergent Larval Capelin Mallotus Villosus. 11
MARINE ECOLOGY - PROGRESS SERIES Vol. 41: 219-229. 1987 Published December 14 Mar. Ecol. Prog. Ser. Short-term mortality in post-emergent larval capelin Mallotus villosus. 11. Importance of food and predator density, and density-dependence* ' Department of Biology, Dalhousie University, Halifax, Nova Scotia B3H 451, Canada Department of Biology, McGill University, 1205 Avenue Docteur Penfield. Montreal, Quebec H3A 1B1. Canada ABSTRACT Analysis of post-emergent larval capehn Mallotus r/lllosus density in a small embayment in eastern Newfoundland durlng 1981 to 1983 demonstrated that as samphng scales approached those relevant to the larvae (ca 2 h 200 to 400 m) the degree of positive association between mlcrozooplank- ton and larvae decreased Forty sequential estimates of post-emergent larval mortality were not slgnlflcantly correlated wlth either the density of edible microzooplankton (90 to 130 um size-classes) the density of potential predators (chaetognaths and lellyflsh) or the density of other macrozooplankton Correlations wlth potential predator densities whlle insignificant, were positive We suggest that posltive correlations between predator denslty and mortallty are more llkely to occur when analysing fleld data that 1s collected near-instantaneously because of the near-instantaneous effect predators have on population losses relative to the longer-term cumulative effects that result from food hmltation (starvation) There was no significant relation between larval density and larval mortallty and the hypothesis of density-dependent short-term mortality in post-emergent larvae was rejected INTRODUCTION 1977, Leggett 1977, Nelson et al. 1977, Bakun & Parish 1980, Bailey 1981, Crecco & Savoy 1984, Leggett et al. In his expansion of Hjort's (1914, 1926) larval mortal- 1984, Sinclair et al. -

Short Communication Captures of Swordfish Xiphias Gladius Linnaeus
Journal of Applied Ichthyology J. Appl. Ichthyol. 26 (2010), 126–127 Received: March 20, 2009 Ó 2010 The Authors Accepted: May 6, 2009 Journal compilation Ó 2010 Blackwell Verlag, Berlin doi: 10.1111/j.1439-0426.2009.01342.x ISSN 0175–8659 Short communication Captures of swordfish Xiphias gladius Linnaeus 1758 and loggerhead sea turtles Caretta caretta (Linnaeus 1758) associated with different bait combinations in the Western Mediterranean surface longline fishery By J. C. Ba´ez1,2, R. Real2, D. Macı´as1, J. M. de la Serna1, J. J. Bellido2,3 and J. A. Camin˜ as1,4 1Instituto Espan˜ol de Oceanografı´a (IEO), Centro Oceanogra´fico de Ma´laga, Ma´laga, Spain; 2Departamento de Biologı´a Animal, Universidad de Ma´laga, Ma´laga, Spain; 3Aula del Mar de Ma´laga, Ma´laga, Spain; 4COPEMED, Oficina de Ma´laga, Ma´laga, Spain Fisheries worldwide use longline surface driftnets mainly for the 18 combinations included fish-only bait, and eight were for targeting swordfish Xiphias gladius Linnaeus, 1758, billfishes a combination of squid ⁄ fish bait. and sharks. Sea turtles Caretta caretta can be incidentally The number of turtles captured using fish-only bait was captured in these surface longlines, considered to be one of the significantly lower than when using squid ⁄ fish baits (per set most dangerous types of fishing gear for sea turtles (Camin˜ as v2 = 238.33; P < 0.0001; per 1000 hooks v2 = 237.296; et al., 2006; Lewison and Crowder, 2007). Thus, there is P < 0.0001) (Table 2). However, there was a significant increasing global interest in strategies to reduce their fishery- reduction in target species catch efficiency when using related incidental catch.