What Do the Structures of GCN5-Containing Complexes Teach Us About Their Function? Dominique Helmlinger, Gábor Papai, Didier Devys, Làszlo Tora
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

SF3B3) and Sin3a Associated Protein 130 (SAP130
cells Communication Ambiguity about Splicing Factor 3b Subunit 3 (SF3B3) and Sin3A Associated Protein 130 (SAP130) Paula I. Metselaar 1,* , Celine Hos 1, Olaf Welting 1, Jos A. Bosch 2,3, Aletta D. Kraneveld 4 , Wouter J. de Jonge 1 and Anje A. Te Velde 1 1 Tytgat Institute for Liver and Intestinal Research, AGEM, Amsterdam UMC, University of Amsterdam, 1105BK Amsterdam, The Netherlands; [email protected] (C.H.); [email protected] (O.W.); [email protected] (W.J.d.J.); [email protected] (A.A.T.V.) 2 Department of Psychology, University of Amsterdam, 1018WS Amsterdam, The Netherlands; [email protected] 3 Department of Medical Psychology, Amsterdam UMC, University of Amsterdam, 1001NK Amsterdam, The Netherlands 4 Division of Pharmacology, Utrecht Institute for Pharmaceutical Sciences, Faculty of Science, Utrecht University, 3584CG Utrecht, The Netherlands; [email protected] * Correspondence: [email protected] Abstract: In 2020, three articles were published on a protein that can activate the immune system by binding to macrophage-inducible C-type lectin receptor (Mincle). In the articles, the protein was referred to as ‘SAP130, a subunit of the histone deacetylase complex.’ However, the Mincle ligand the authors aimed to investigate is splicing factor 3b subunit 3 (SF3B3). This splicing factor is unrelated to SAP130 (Sin3A associated protein 130, a subunit of the histone deacetylase-dependent Sin3A corepressor complex). The conclusions in the three articles were formulated for SF3B3, Citation: Metselaar, P.I.; Hos, C.; while the researchers used qPCR primers and antibodies against SAP130. -

Gene Regulation and Speciation in House Mice
Downloaded from genome.cshlp.org on September 26, 2021 - Published by Cold Spring Harbor Laboratory Press Research Gene regulation and speciation in house mice Katya L. Mack,1 Polly Campbell,2 and Michael W. Nachman1 1Museum of Vertebrate Zoology and Department of Integrative Biology, University of California, Berkeley, California 94720-3160, USA; 2Department of Integrative Biology, Oklahoma State University, Stillwater, Oklahoma 74078, USA One approach to understanding the process of speciation is to characterize the genetic architecture of post-zygotic isolation. As gene regulation requires interactions between loci, negative epistatic interactions between divergent regulatory elements might underlie hybrid incompatibilities and contribute to reproductive isolation. Here, we take advantage of a cross between house mouse subspecies, where hybrid dysfunction is largely unidirectional, to test several key predictions about regulatory divergence and reproductive isolation. Regulatory divergence between Mus musculus musculus and M. m. domesticus was charac- terized by studying allele-specific expression in fertile hybrid males using mRNA-sequencing of whole testes. We found ex- tensive regulatory divergence between M. m. musculus and M. m. domesticus, largely attributable to cis-regulatory changes. When both cis and trans changes occurred, they were observed in opposition much more often than expected under a neutral model, providing strong evidence of widespread compensatory evolution. We also found evidence for lineage-specific positive se- lection on a subset of genes related to transcriptional regulation. Comparisons of fertile and sterile hybrid males identified a set of genes that were uniquely misexpressed in sterile individuals. Lastly, we discovered a nonrandom association between these genes and genes showing evidence of compensatory evolution, consistent with the idea that regulatory interactions might contribute to Dobzhansky-Muller incompatibilities and be important in speciation. -

SF3B2-Mediated RNA Splicing Drives Human Prostate Cancer Progression
Published OnlineFirst August 20, 2019; DOI: 10.1158/0008-5472.CAN-18-3965 Cancer Molecular Cell Biology Research SF3B2-Mediated RNA Splicing Drives Human Prostate Cancer Progression Norihiko Kawamura1,2, Keisuke Nimura1, Kotaro Saga1, Airi Ishibashi1, Koji Kitamura1,3, Hiromichi Nagano1, Yusuke Yoshikawa4, Kyoso Ishida1,5, Norio Nonomura2, Mitsuhiro Arisawa4, Jun Luo6, and Yasufumi Kaneda1 Abstract Androgen receptor splice variant-7 (AR-V7) is a General RNA splicing SF3B2 complex-mediated alternative RNA splicing constitutively active AR variant implicated in U2 castration-resistant prostate cancers. Here, we show U2 snRNA that the RNA splicing factor SF3B2, identified by 3’ 3’ in silico and CRISPR/Cas9 analyses, is a critical 5’ 3’ splice site 5’ SF3B7 AR-V7 5’ A U2AF2 AGA Exon ? determinant of expression and is correlated SF3B6(p14) SF3B4 SF3B1 SF3B4 SF3B1 with aggressive cancer phenotypes. Transcriptome SF3B5 SF3B2 SF3B3 SF3B2 SF3B3 and PAR-CLIP analyses revealed that SF3B2 con- SF3A3 SF3B2 complex SF3A3 SF3A1 SF3A1 SF3b complex trols the splicing of target genes, including AR, to AR pre-mRNA drive aggressive phenotypes. SF3B2-mediated CE3 aggressive phenotypes in vivo were reversed by AR-V7 mRNA AR mRNA AR-V7 knockout. Pladienolide B, an inhibitor of CE3 a splicing modulator of the SF3b complex, sup- Drive malignancy pressed the growth of tumors addicted to high While the SF3b complex is critical for general RNA splicing, SF3B2 promotes inclusion of the target exon through recognizing a specific RNA motif. SF3B2 expression. These findings support the idea © 2019 American Association for Cancer Research that alteration of the splicing pattern by high SF3B2 expression is one mechanism underlying prostate cancer progression and therapeutic resistance. -

Insights Into Regulation of Human RAD51 Nucleoprotein Filament Activity During
Insights into Regulation of Human RAD51 Nucleoprotein Filament Activity During Homologous Recombination Dissertation Presented in Partial Fulfillment of the Requirements for the Degree Doctor of Philosophy in the Graduate School of The Ohio State University By Ravindra Bandara Amunugama, B.S. Biophysics Graduate Program The Ohio State University 2011 Dissertation Committee: Richard Fishel PhD, Advisor Jeffrey Parvin MD PhD Charles Bell PhD Michael Poirier PhD Copyright by Ravindra Bandara Amunugama 2011 ABSTRACT Homologous recombination (HR) is a mechanistically conserved pathway that occurs during meiosis and following the formation of DNA double strand breaks (DSBs) induced by exogenous stresses such as ionization radiation. HR is also involved in restoring replication when replication forks have stalled or collapsed. Defective recombination machinery leads to chromosomal instability and predisposition to tumorigenesis. However, unregulated HR repair system also leads to similar outcomes. Fortunately, eukaryotes have evolved elegant HR repair machinery with multiple mediators and regulatory inputs that largely ensures an appropriate outcome. A fundamental step in HR is the homology search and strand exchange catalyzed by the RAD51 recombinase. This process requires the formation of a nucleoprotein filament (NPF) on single-strand DNA (ssDNA). In Chapter 2 of this dissertation I describe work on identification of two residues of human RAD51 (HsRAD51) subunit interface, F129 in the Walker A box and H294 of the L2 ssDNA binding region that are essential residues for salt-induced recombinase activity. Mutation of F129 or H294 leads to loss or reduced DNA induced ATPase activity and formation of a non-functional NPF that eliminates recombinase activity. DNA binding studies indicate that these residues may be essential for sensing the ATP nucleotide for a functional NPF formation. -

Genome-Wide DNA Methylation Analysis Reveals Molecular Subtypes of Pancreatic Cancer
www.impactjournals.com/oncotarget/ Oncotarget, 2017, Vol. 8, (No. 17), pp: 28990-29012 Research Paper Genome-wide DNA methylation analysis reveals molecular subtypes of pancreatic cancer Nitish Kumar Mishra1 and Chittibabu Guda1,2,3,4 1Department of Genetics, Cell Biology and Anatomy, University of Nebraska Medical Center, Omaha, NE, 68198, USA 2Bioinformatics and Systems Biology Core, University of Nebraska Medical Center, Omaha, NE, 68198, USA 3Department of Biochemistry and Molecular Biology, University of Nebraska Medical Center, Omaha, NE, 68198, USA 4Fred and Pamela Buffet Cancer Center, University of Nebraska Medical Center, Omaha, NE, 68198, USA Correspondence to: Chittibabu Guda, email: [email protected] Keywords: TCGA, pancreatic cancer, differential methylation, integrative analysis, molecular subtypes Received: October 20, 2016 Accepted: February 12, 2017 Published: March 07, 2017 Copyright: Mishra et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC-BY), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. ABSTRACT Pancreatic cancer (PC) is the fourth leading cause of cancer deaths in the United States with a five-year patient survival rate of only 6%. Early detection and treatment of this disease is hampered due to lack of reliable diagnostic and prognostic markers. Recent studies have shown that dynamic changes in the global DNA methylation and gene expression patterns play key roles in the PC development; hence, provide valuable insights for better understanding the initiation and progression of PC. In the current study, we used DNA methylation, gene expression, copy number, mutational and clinical data from pancreatic patients. -

Identification of a Proteomic Signature of Senescence in Primary Human
bioRxiv preprint doi: https://doi.org/10.1101/2020.09.22.309351; this version posted January 12, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Identification of a Proteomic Signature of Senescence in Primary Human Mammary Epithelial Cells Alireza Delfarah1†, DongQing Zheng1, Jesse Yang1 and Nicholas A. Graham1,2,3 1 Mork Family Department of Chemical Engineering and Materials Science, 2 Norris Comprehensive Cancer Center, 3 Leonard Davis School of Gerontology, University of Southern California, Los Angeles, CA 90089 †Current address, Department of Genetics, Stanford University, Palo Alto, CA 94305 Running title: Proteomic profiling of primary HMEC senescence Keywords: replicative senescence, aging, mammary epithelial cell, proteomics, data integration, transcriptomics Corresponding author: Nicholas A. Graham, University of Southern California, 3710 McClintock Ave., RTH 509, Los Angeles, CA 90089. Phone: 213-240-0449; E-mail: [email protected] bioRxiv preprint doi: https://doi.org/10.1101/2020.09.22.309351; this version posted January 12, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Proteomic profiling of primary HMEC senescence Abstract Senescence is a permanent cell cycle arrest that occurs in response to cellular stress. Because senescent cells promote age-related disease, there has been considerable interest in defining the proteomic alterations in senescent cells. Because senescence differs greatly depending on cell type and senescence inducer, continued progress in the characterization of senescent cells is needed. Here, we analyzed primary human mammary epithelial cells (HMECs), a model system for aging, using mass spectrometry-based proteomics. -

Open Data for Differential Network Analysis in Glioma
International Journal of Molecular Sciences Article Open Data for Differential Network Analysis in Glioma , Claire Jean-Quartier * y , Fleur Jeanquartier y and Andreas Holzinger Holzinger Group HCI-KDD, Institute for Medical Informatics, Statistics and Documentation, Medical University Graz, Auenbruggerplatz 2/V, 8036 Graz, Austria; [email protected] (F.J.); [email protected] (A.H.) * Correspondence: [email protected] These authors contributed equally to this work. y Received: 27 October 2019; Accepted: 3 January 2020; Published: 15 January 2020 Abstract: The complexity of cancer diseases demands bioinformatic techniques and translational research based on big data and personalized medicine. Open data enables researchers to accelerate cancer studies, save resources and foster collaboration. Several tools and programming approaches are available for analyzing data, including annotation, clustering, comparison and extrapolation, merging, enrichment, functional association and statistics. We exploit openly available data via cancer gene expression analysis, we apply refinement as well as enrichment analysis via gene ontology and conclude with graph-based visualization of involved protein interaction networks as a basis for signaling. The different databases allowed for the construction of huge networks or specified ones consisting of high-confidence interactions only. Several genes associated to glioma were isolated via a network analysis from top hub nodes as well as from an outlier analysis. The latter approach highlights a mitogen-activated protein kinase next to a member of histondeacetylases and a protein phosphatase as genes uncommonly associated with glioma. Cluster analysis from top hub nodes lists several identified glioma-associated gene products to function within protein complexes, including epidermal growth factors as well as cell cycle proteins or RAS proto-oncogenes. -

Mutations in Splicing Factor Genes in Myeloid Malignancies: Significance and Impact on Clinical Features
cancers Review Mutations in Splicing Factor Genes in Myeloid Malignancies: Significance and Impact on Clinical Features Valeria Visconte 1, Megan O. Nakashima 2 and Heesun J. Rogers 2,* 1 Department of Translational Hematology and Oncology Research, Taussig Cancer Institute, Cleveland Clinic, Cleveland, OH 44195, USA; [email protected] 2 Department of Laboratory Medicine, Cleveland Clinic, Cleveland, OH 44195, USA; [email protected] * Correspondence: [email protected]; Tel.: +1-216-445-2719 Received: 15 October 2019; Accepted: 19 November 2019; Published: 22 November 2019 Abstract: Components of the pre-messenger RNA splicing machinery are frequently mutated in myeloid malignancies. Mutations in LUC7L2, PRPF8, SF3B1, SRSF2, U2AF1, and ZRSR2 genes occur at various frequencies ranging between 40% and 85% in different subtypes of myelodysplastic syndrome (MDS) and 5% and 10% of acute myeloid leukemia (AML) and myeloproliferative neoplasms (MPNs). In some instances, splicing factor (SF) mutations have provided diagnostic utility and information on clinical outcomes as exemplified by SF3B1 mutations associated with increased ring sideroblasts (RS) in MDS-RS or MDS/MPN-RS with thrombocytosis. SF3B1 mutations are associated with better survival outcomes, while SRSF2 mutations are associated with a shorter survival time and increased AML progression, and U2AF1 mutations with a lower remission rate and shorter survival time. Beside the presence of mutations, transcriptomics technologies have shown that one third of genes in AML patients are differentially expressed, leading to altered transcript stability, interruption of protein function, and improper translation compared to those of healthy individuals. The detection of SF mutations demonstrates the importance of splicing abnormalities in the hematopoiesis of MDS and AML patients given the fact that abnormal splicing regulates the function of several transcriptional factors (PU.1, RUNX1, etc.) crucial in hematopoietic function. -
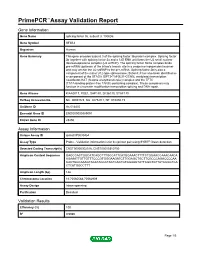
Primepcr™Assay Validation Report
PrimePCR™Assay Validation Report Gene Information Gene Name splicing factor 3b, subunit 3, 130kDa Gene Symbol SF3B3 Organism Human Gene Summary This gene encodes subunit 3 of the splicing factor 3b protein complex. Splicing factor 3b together with splicing factor 3a and a 12S RNA unit forms the U2 small nuclear ribonucleoproteins complex (U2 snRNP). The splicing factor 3b/3a complex binds pre-mRNA upstream of the intron's branch site in a sequence independent manner and may anchor the U2 snRNP to the pre-mRNA. Splicing factor 3b is also a component of the minor U12-type spliceosome. Subunit 3 has also been identified as a component of the STAGA (SPT3-TAF(II)31-GCN5L acetylase) transcription coactivator-HAT (histone acetyltransferase) complex and the TFTC (TATA-binding-protein-free TAF(II)-containing complex). These complexes may function in chromatin modification transcription splicing and DNA repair. Gene Aliases KIAA0017, RSE1, SAP130, SF3b130, STAF130 RefSeq Accession No. NC_000016.9, NG_027529.1, NT_010498.15 UniGene ID Hs.514435 Ensembl Gene ID ENSG00000189091 Entrez Gene ID 23450 Assay Information Unique Assay ID qHsaCIP0030454 Assay Type Probe - Validation information is for the primer pair using SYBR® Green detection Detected Coding Transcript(s) ENST00000302516, ENST00000310750 Amplicon Context Sequence GAGCCACTGGCATCAGCTTTGCCATTCATGGAAACTTTTCTGGAACCAAACAACA AGAAATTGTTGTTTCCCGTGGGAAGATCTTGGAGCTGCTTCGCCCAGACCCCAA CACTGGCAAAGTACATACCCTACTCACTGTGGAAGTATTCGGTGTTATCCGGTCA CTCATGGCCTTT Amplicon Length (bp) 146 Chromosome Location 16:70560588-70562909 Assay Design Intron-spanning Purification Desalted Validation Results Efficiency (%) 100 R2 0.9986 Page 1/5 PrimePCR™Assay Validation Report cDNA Cq 18.72 cDNA Tm (Celsius) 83 gDNA Cq Specificity (%) 100 Information to assist with data interpretation is provided at the end of this report. -

A High-Throughput Approach to Uncover Novel Roles of APOBEC2, a Functional Orphan of the AID/APOBEC Family
Rockefeller University Digital Commons @ RU Student Theses and Dissertations 2018 A High-Throughput Approach to Uncover Novel Roles of APOBEC2, a Functional Orphan of the AID/APOBEC Family Linda Molla Follow this and additional works at: https://digitalcommons.rockefeller.edu/ student_theses_and_dissertations Part of the Life Sciences Commons A HIGH-THROUGHPUT APPROACH TO UNCOVER NOVEL ROLES OF APOBEC2, A FUNCTIONAL ORPHAN OF THE AID/APOBEC FAMILY A Thesis Presented to the Faculty of The Rockefeller University in Partial Fulfillment of the Requirements for the degree of Doctor of Philosophy by Linda Molla June 2018 © Copyright by Linda Molla 2018 A HIGH-THROUGHPUT APPROACH TO UNCOVER NOVEL ROLES OF APOBEC2, A FUNCTIONAL ORPHAN OF THE AID/APOBEC FAMILY Linda Molla, Ph.D. The Rockefeller University 2018 APOBEC2 is a member of the AID/APOBEC cytidine deaminase family of proteins. Unlike most of AID/APOBEC, however, APOBEC2’s function remains elusive. Previous research has implicated APOBEC2 in diverse organisms and cellular processes such as muscle biology (in Mus musculus), regeneration (in Danio rerio), and development (in Xenopus laevis). APOBEC2 has also been implicated in cancer. However the enzymatic activity, substrate or physiological target(s) of APOBEC2 are unknown. For this thesis, I have combined Next Generation Sequencing (NGS) techniques with state-of-the-art molecular biology to determine the physiological targets of APOBEC2. Using a cell culture muscle differentiation system, and RNA sequencing (RNA-Seq) by polyA capture, I demonstrated that unlike the AID/APOBEC family member APOBEC1, APOBEC2 is not an RNA editor. Using the same system combined with enhanced Reduced Representation Bisulfite Sequencing (eRRBS) analyses I showed that, unlike the AID/APOBEC family member AID, APOBEC2 does not act as a 5-methyl-C deaminase. -

NRF1) Coordinates Changes in the Transcriptional and Chromatin Landscape Affecting Development and Progression of Invasive Breast Cancer
Florida International University FIU Digital Commons FIU Electronic Theses and Dissertations University Graduate School 11-7-2018 Decipher Mechanisms by which Nuclear Respiratory Factor One (NRF1) Coordinates Changes in the Transcriptional and Chromatin Landscape Affecting Development and Progression of Invasive Breast Cancer Jairo Ramos [email protected] Follow this and additional works at: https://digitalcommons.fiu.edu/etd Part of the Clinical Epidemiology Commons Recommended Citation Ramos, Jairo, "Decipher Mechanisms by which Nuclear Respiratory Factor One (NRF1) Coordinates Changes in the Transcriptional and Chromatin Landscape Affecting Development and Progression of Invasive Breast Cancer" (2018). FIU Electronic Theses and Dissertations. 3872. https://digitalcommons.fiu.edu/etd/3872 This work is brought to you for free and open access by the University Graduate School at FIU Digital Commons. It has been accepted for inclusion in FIU Electronic Theses and Dissertations by an authorized administrator of FIU Digital Commons. For more information, please contact [email protected]. FLORIDA INTERNATIONAL UNIVERSITY Miami, Florida DECIPHER MECHANISMS BY WHICH NUCLEAR RESPIRATORY FACTOR ONE (NRF1) COORDINATES CHANGES IN THE TRANSCRIPTIONAL AND CHROMATIN LANDSCAPE AFFECTING DEVELOPMENT AND PROGRESSION OF INVASIVE BREAST CANCER A dissertation submitted in partial fulfillment of the requirements for the degree of DOCTOR OF PHILOSOPHY in PUBLIC HEALTH by Jairo Ramos 2018 To: Dean Tomás R. Guilarte Robert Stempel College of Public Health and Social Work This dissertation, Written by Jairo Ramos, and entitled Decipher Mechanisms by Which Nuclear Respiratory Factor One (NRF1) Coordinates Changes in the Transcriptional and Chromatin Landscape Affecting Development and Progression of Invasive Breast Cancer, having been approved in respect to style and intellectual content, is referred to you for judgment. -

The Biological Function and Clinical Significance of SF3B1 Mutations In
Zhou et al. Biomarker Research (2020) 8:38 https://doi.org/10.1186/s40364-020-00220-5 REVIEW Open Access The biological function and clinical significance of SF3B1 mutations in cancer Zhixia Zhou1* , Qi Gong2, Yin Wang1, Mengkun Li1, Lu Wang1, Hongfei Ding1 and Peifeng Li1* Abstract Spliceosome mutations have become the most interesting mutations detected in human cancer in recent years. The spliceosome, a large, dynamic multimegadalton small nuclear ribonucleoprotein composed of small nuclear RNAs associated with proteins, is responsible for removing introns from precursor mRNA (premRNA) and generating mature, spliced mRNAs. SF3B1 is the largest subunit of the spliceosome factor 3b (SF3B) complex, which is a core component of spliceosomes. Recurrent somatic mutations in SF3B1 have been detected in human cancers, including hematological malignancies and solid tumors, and indicated to be related to patient prognosis. This review summarizes the research progress of SF3B1 mutations in cancer, including SF3B1 mutations in the HEAT domain, the multiple roles and aberrant splicing events of SF3B1 mutations in the pathogenesis of tumors, and changes in mutated cancer cells regarding sensitivity to SF3B small-molecule inhibitors. In addition, the potential of SF3B1 or its mutations to serve as biomarkers or therapeutic targets in cancer is discussed. The accumulated knowledge about SF3B1 mutations in cancer provides critical insight into the integral role the SF3B1 protein plays in mRNA splicing and suggests new targets for anticancer therapy. Keywords: SF3B1, Mutation, Cancer, RNA splicing Background human diseases, including cancer [5, 6]. Indeed, genome- Of the 3.3 billion base pairs of haploid DNA in the human wide studies have revealed more than 15,000 tumor- genome, approximately 20,000 protein-coding genes have associated splice variants in a wide variety of cancers [7, 8].