The Developing Mouse Coronal Suture at Single-Cell Resolution
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Coronal and Lambdoid Suture Evolution Following Total Vault Remodeling for Scaphocephaly
NEUROSURGICAL FOCUS Neurosurg Focus 50 (4):E4, 2021 Coronal and lambdoid suture evolution following total vault remodeling for scaphocephaly Pierre-Aurélien Beuriat, MD, PhD,1,2,4 Alexandru Szathmari, MD, PhD,1,2 Julie Chauvel-Picard, MD,1,3,4 Arnaud Gleizal, MD, PhD,1,3,4 Christian Paulus, MD,1,3 Carmine Mottolese, MD, PhD,1,2 and Federico Di Rocco, MD, PhD1,2,4 1French Referral Center for Craniosynostosis; Departments of 2Pediatric Neurosurgery and 3Pediatric Maxillo-Facial Surgery, Hôpital Femme Mère Enfant; and 4Université de Lyon, France OBJECTIVE Different types of surgical procedures are utilized to treat craniosynostosis. In most procedures, the fused suture is removed. There are only a few reports on the evolution of sutures after surgical correction of craniosynostosis. To date, no published study describes neosuture formation after total cranial vault remodeling. The objective of this study was to understand the evolution of the cranial bones in the area of coronal and lambdoid sutures that were removed for complete vault remodeling in patients with sagittal craniosynostosis. In particular, the investigation aimed to confirm the possibility of neosuture formation. METHODS CT images of the skulls of children who underwent operations for scaphocephaly at the Hôpital Femme Mère Enfant, Lyon University Hospital, Lyon, France, from 2004 to 2014 were retrospectively reviewed. Inclusion crite- ria were diagnosis of isolated sagittal synostosis, age between 4 and 18 months at surgery, and availability of reliable postoperative CT images obtained at a minimum of 1 year after surgical correction. Twenty-six boys and 11 girls were included, with a mean age at surgery of 231.6 days (range 126–449 days). -

Frontosphenoidal Synostosis: a Rare Cause of Unilateral Anterior Plagiocephaly
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by RERO DOC Digital Library Childs Nerv Syst (2007) 23:1431–1438 DOI 10.1007/s00381-007-0469-4 ORIGINAL PAPER Frontosphenoidal synostosis: a rare cause of unilateral anterior plagiocephaly Sandrine de Ribaupierre & Alain Czorny & Brigitte Pittet & Bertrand Jacques & Benedict Rilliet Received: 30 March 2007 /Published online: 22 September 2007 # Springer-Verlag 2007 Abstract Conclusion Frontosphenoidal synostosis must be searched Introduction When a child walks in the clinic with a in the absence of a coronal synostosis in a child with unilateral frontal flattening, it is usually associated in our anterior unilateral plagiocephaly, and treated surgically. minds with unilateral coronal synostosis. While the latter might be the most common cause of anterior plagiocephaly, Keywords Craniosynostosis . Pediatric neurosurgery. it is not the only one. A patent coronal suture will force us Anterior plagiocephaly to consider other etiologies, such as deformational plagio- cephaly, or synostosis of another suture. To understand the mechanisms underlying this malformation, the development Introduction and growth of the skull base must be considered. Materials and methods There have been few reports in the Harmonious cranial growth is dependent on patent sutures, literature of isolated frontosphenoidal suture fusion, and and any craniosynostosis might lead to an asymmetrical we would like to report a series of five cases, as the shape of the skull. The anterior skull base is formed of recognition of this entity is important for its treatment. different bones, connected by sutures, fusing at different ages. The frontosphenoidal suture extends from the end of Presented at the Consensus Conference on Pediatric Neurosurgery, the frontoparietal suture, anteriorly and inferiorly in the Rome, 1–2 December 2006. -

Cleidocranial Dysplasia- a Case Report
Case Report Cleidocranial Dysplasia- A Case Report Akhilanand Chaurasia Department of Oral Medicine & Radiology, Faculty of Dental Sciences, King George Medical University, Lucknow E-mail: [email protected] ABSTRACT Cleidocranial dysplasia is a rare congenital disease. It is characterized by autosomal dominant inheritance pattern which is caused due to mutations in the Cbfa1 gene (Runx2) located on chromosome 6p21. It primarily affects bones which are formedby intra-membranous ossification and have equal sex distribution. It is also known as Marie and Sainton disease, Mutational dysostosis and cleidocranialdysostosis. The skeletal deformities of cleidocranial dysplasia are characterized by partial or complete absence of clavicles, late closure of the fontanels, presence of open skull sutures and multiple wormian bones. This rare syndrome is of utmost importance in dentistry due to presence of multiple supernumerary teeth, facial bones deformities and deranged eruption patterns. We are reporting a classical case of cleidocranial dysplasia in 20 year old patient. Keywords: Cleidocranial dysplasia, Marie and Sainton disease, Mutational dysostosis, Cleidocranialdysostosis, Autosomal dominant Access this article online mandibular symphysis are less common findings of Quick Response cleidocranial dysostosis1,10,11. Code: Website: Dental findings in cleidocranialdysostosis are www.innovativepublication.com characterized by a decreased eruptive force of both primary and permanent dentition, prolonged retention of primary teeth12 and an increase in odontogenesis DOI: 10.5958/2395-6194.2015.00009.0 leading to an excessive number of supernumerary teeth13. The clinical findings of cleidocranialdysostosis although present at birth are often either missed or INTRODUCTION diagnosed at a much later time. Cleidocranialdysostosis Cleidocranialdysostosis is a rare congenital defect may be identified by family history, excessive mobility primarily affecting bones which undergo intra- of shoulders and radiographic pathognomonic findings membranous ossification i.e. -

Cranial Suture Mesenchymal Stem Cells: Insights and Advances
biomolecules Review Cranial Suture Mesenchymal Stem Cells: Insights and Advances Bo Li 1, Yigan Wang 1, Yi Fan 2, Takehito Ouchi 3 , Zhihe Zhao 1,* and Longjiang Li 4,* 1 State Key Laboratory of Oral Diseases, National Clinical Research Center for Oral Diseases, Department of Orthodontics, West China Hospital of Stomatology, Sichuan University, Chengdu 610041, China; [email protected] (B.L.); [email protected] (Y.W.) 2 State Key Laboratory of Oral Diseases, National Clinical Research Center for Oral Diseases, Department of Cariology and Endodontics, West China Hospital of Stomatology, Sichuan University, Chengdu 610041, China; [email protected] 3 Department of Physiology, Tokyo Dental College, Tokyo 1010061, Japan; [email protected] 4 State Key Laboratory of Oral Diseases, National Clinical Research Center for Oral Diseases, Department of Head and Neck Oncology, West China Hospital of Stomatology, Sichuan University, Chengdu 610041, China * Correspondence: [email protected] (Z.Z.); [email protected] (L.L.) Abstract: The cranial bones constitute the protective structures of the skull, which surround and protect the brain. Due to the limited repair capacity, the reconstruction and regeneration of skull defects are considered as an unmet clinical need and challenge. Previously, it has been proposed that the periosteum and dura mater provide reparative progenitors for cranial bones homeostasis and injury repair. In addition, it has also been speculated that the cranial mesenchymal stem cells reside in the perivascular niche of the diploe, namely, the soft spongy cancellous bone between the interior and exterior layers of cortical bone of the skull, which resembles the skeletal stem cells’ distribution pattern of the long bone within the bone marrow. -

Ectocranial Suture Closure in Pan Troglodytes and Gorilla Gorilla: Pattern and Phylogeny James Cray Jr.,1* Richard S
AMERICAN JOURNAL OF PHYSICAL ANTHROPOLOGY 136:394–399 (2008) Ectocranial Suture Closure in Pan troglodytes and Gorilla gorilla: Pattern and Phylogeny James Cray Jr.,1* Richard S. Meindl,2 Chet C. Sherwood,3 and C. Owen Lovejoy2 1Department of Anthropology, University of Pittsburgh, Pittsburgh, PA 15260 2Department of Anthropology and Division of Biomedical Sciences, Kent State University, Kent, OH 44242 3Department of Anthropology, The George Washington University, Washington, DC 20052 KEY WORDS cranial suture; synostosis; variation; phylogeny; Guttman analysis ABSTRACT The order in which ectocranial sutures than either does with G. gorilla, we hypothesized that this undergo fusion displays species-specific variation among phylogenetic relationship would be reflected in the suture primates. However, the precise relationship between suture closure patterns of these three taxa. Results indicated that closure and phylogenetic affinities is poorly understood. In while all three species do share a similar lateral-anterior this study, we used Guttman Scaling to determine if the closure pattern, G. gorilla exhibits a unique vault pattern, modal progression of suture closure differs among Homo which, unlike humans and P. troglodyte s, follows a strong sapiens, Pan troglodytes,andGorilla gorilla.BecauseDNA posterior-to-anterior gradient. P. troglodytes is therefore sequence homologies strongly suggest that P. tr og lodytes more like Homo sapiens in suture synostosis. Am J Phys and Homo sapiens share a more recent common ancestor Anthropol 136:394–399, 2008. VC 2008 Wiley-Liss, Inc. The biological basis of suture synostosis is currently Morriss-Kay et al. (2001) found that maintenance of pro- poorly understood, but appears to be influenced by a liferating osteogenic stem cells at the margins of mem- combination of vascular, hormonal, genetic, mechanical, brane bones forming the coronal suture requires FGF and local factors (see review in Cohen, 1993). -

Gen Anat-Joints
JOINTS Joint is a junction between two or more bones Classification •Functional Based on the range and type of movement they permit •Structural On the basis of their anatomic structure Functional Classification • Synarthrosis No movement e.g. Fibrous joint • Amphiarthrosis Slight movement e.g. Cartilagenous joint • Diarthrosis Movement present Cavity present Also called as Synovial joint eg.shoulder joint Structural Classification Based on type of connective tissue binding the two adjacent articulating bones Presence or absence of synovial cavity in between the articulating bone • Fibrous • Cartilagenous • Synovial Fibrous Joint Bones are connected to each other by fibrous (connective ) tissue No movement No synovial cavity • Suture • Syndesmosis • Gomphosis Sutural Joints • A thin layer of dens fibrous tissue binds the adjacent bones • These appear between the bones which ossify in membrane • Present between the bones of skull e.g . coronal suture, sagittal suture • Schindylesis: – rigid bone fits in to a groove on a neighbouring bone e.g. Vomer and sphenoid Gomphosis • Peg and socket variety • Cone shaped root of tooth fits in to a socket of jaw • Immovable • Root is attached to the socket by fibrous tissue (periodontal ligament). Syndesmosis • Bony surfaces are bound together by interosseous ligament or membrane • Membrane permits slight movement • Functionally classified as amphiarthrosis e.g. inferior tibiofibular joint Cartilaginous joint • Bones are held together by cartilage • Absence of synovial cavity . Synchondrosis . Symphysis Synchondrosis • Primary cartilaginous joint • Connecting material between two bones is hyaline cartilage • Temporary joint • Immovable joint • After a certain age cartilage is replaced by bone (synostosis) • e.g. Epiphyseal plate connecting epiphysis and diphysis of a long bone, joint between basi-occiput and basi-sphenoid Symphysis • Secondary cartilaginous joint (fibrocartilaginous joint) • Permanent joint • Occur in median plane of the body • Slightly movable • e.g. -

Modelling Suture Ossification: a View from the Cranial Capsule
University of Tennessee, Knoxville TRACE: Tennessee Research and Creative Exchange Masters Theses Graduate School 8-1992 Modelling Suture Ossification: A View from the Cranial Capsule Hugh Bryson Matternes University of Tennessee, Knoxville Follow this and additional works at: https://trace.tennessee.edu/utk_gradthes Part of the Anthropology Commons Recommended Citation Matternes, Hugh Bryson, "Modelling Suture Ossification: A View from the Cranial Capsule. " Master's Thesis, University of Tennessee, 1992. https://trace.tennessee.edu/utk_gradthes/4192 This Thesis is brought to you for free and open access by the Graduate School at TRACE: Tennessee Research and Creative Exchange. It has been accepted for inclusion in Masters Theses by an authorized administrator of TRACE: Tennessee Research and Creative Exchange. For more information, please contact [email protected]. To the Graduate Council: I am submitting herewith a thesis written by Hugh Bryson Matternes entitled "Modelling Suture Ossification: A View from the Cranial Capsule." I have examined the final electronic copy of this thesis for form and content and recommend that it be accepted in partial fulfillment of the requirements for the degree of Master of Arts, with a major in Anthropology. Richard L. Jantz, Major Professor We have read this thesis and recommend its acceptance: Lyle W. Konigsberg, William M. Bass Accepted for the Council: Carolyn R. Hodges Vice Provost and Dean of the Graduate School (Original signatures are on file with official studentecor r ds.) To the Graduate Council: -

Cranial Sutures & Funny Shaped Heads: Radiological Diagnosis
Objectives • The objectives of this presentation are to: – Review the imaging features of normal cranial sutures – Identify the characteristics of abnormal skull shape on imaging – Review the characteristics of the most common non- syndromic and syndromic causes of craniosynostosis Anatomical Review Anatomical Review • The bony plates of the skull communicate at the cranial sutures • The anterior fontanelle occurs where the coronal & metopic sutures meet • The posterior fontanelle occurs where the sagittal & lambdoid sutures meet Anatomical Review • The main cranial sutures & fontanelles include: Metopic Suture Anterior Fontanelle Coronal Sutures Squamosal Sutures Posterior Fontanelle Sagittal Suture Lambdoid Sutures Anatomical Review • Growth of the skull occurs perpendicular to the cranial suture • This is controlled by a complex signalling system including: – Ephrins (mark the suture boundary) – Fibroblast growth factor receptors (FGFR) – Transcription factor TWIST Anatomical Review • The cranial sutures are important for rapid skull growth in-utero & infancy • The cranial sutures can usually be visualised on imaging into late adulthood Normal Radiological Appearances Normal Radiological Appearances • The cranial sutures can be visualised on plain radiographs • Standard views include: – PA – Lateral – Townes PA Skull radiograph Sagittal Suture Left Coronal Suture Right CoronalRight lambdoidSutureSuture Metopic Suture Left lambdoid Suture Townes View Sagittal Suture Left Coronal Suture Right Coronal Suture Right Lambdoid Suture Left -

A Heads up on Craniosynostosis
Volume 1, Issue 1 A HEADS UP ON CRANIOSYNOSTOSIS Andrew Reisner, M.D., William R. Boydston, M.D., Ph.D., Barun Brahma, M.D., Joshua Chern, M.D., Ph.D., David Wrubel, M.D. Craniosynostosis, an early closure of the growth plates of the skull, results in a skull deformity and may result in neurologic compromise. Craniosynostosis is surprisingly common, occurring in one in 2,100 children. It may occur as an isolated abnormality, as a part of a syndrome or secondary to a systemic disorder. Typically, premature closure of a suture results in a characteristic cranial deformity easily recognized by a trained observer. it is usually the misshapen head that brings the child to receive medical attention and mandates treatment. This issue of Neuro Update will present the diagnostic features of the common types of craniosynostosis to facilitate recognition by the primary care provider. The distinguishing features of other conditions commonly confused with craniosynostosis, such as plagiocephaly, benign subdural hygromas of infancy and microcephaly, will be discussed. Treatment options for craniosynostosis will be reserved for a later issue of Neuro Update. Types of Craniosynostosis Sagittal synostosis The sagittal suture runs from the anterior to the posterior fontanella (Figure 1). Like the other sutures, it allows the cranial bones to overlap during birth, facilitating delivery. Subsequently, this suture is the separation between the parietal bones, allowing lateral growth of the midportion of the skull. Early closure of the sagittal suture restricts lateral growth and creates a head that is narrow from ear to ear. However, a single suture synostosis causes deformity of the entire calvarium. -
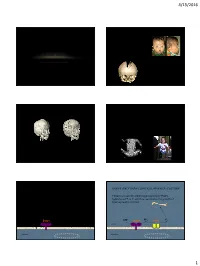
Durussel John -- Bone Mineralization Dependent Craniosynostosis and Craniofacial Shape Abnormalities.Pdf
4/15/2016 Infant with craniosynostosis CRANIOSYNOSTOSIS • Premature fusion of cranial bones ANALYSIS OF CRANIOFACIAL • 1/2500 live births MORPHOLOGY IN A TNAP KNOCKOUT • May involve one or multiple sutures • Isolated (85%) or Syndromic (15%) MOUSE MODEL • Biologic process unknown • Elevated intracranial pressure John DuRussel MS, DDS, MS • Impaired cerebral blood flow • Airway obstruction • Deafness, blindness, seizures • Developmental delay and learning disabilities • Esthetic compromises CURRENT TREAMENT: CRANIAL VAULT REMODELING SURGERY HYPOPHOSPHATASIA (HPP) • Deficiency of Tissue Non-specific Alkaline Phosphatase (TNAP) • Disrupted mineralization of the skeleton and dentition (weak bones and teeth) • Seizures due to poor Vitamin B metabolism • Craniosynostosis in the context of low bone density (40% of patients) • Death (respiratory dysfunction due to weak rib bones) • Excise prematurely fused sutures and correct calvarial deformities • Initial surgery at 3-6 months old to allow for exponential brain growth ENPP1 AND TNAP CONTROL MINERALIZATION ENPP1 AND TNAP CONTROL MINERALIZATION • Ectonucleotide pyrophosphatase/phosphodiesterase-1 • Tissue non-specific alkaline phosphatase (TNAP) (Enpp1) - the primary osteoblastic generator of hydrolyzes PPi to Pi which is essential to the growth of pyrophosphate (PPi) hydroxyapatite crystals. ATP Enpp1 PPi ATP Enpp1 PPi TNAP Pi Matrix Matrix Osteoblast Osteoblast 1 4/15/2016 Qualitative Craniofacial Phenotype of the ENPP1 AND TNAP CONTROL MINERALIZATION TNAP-/- Mouse Model of Infantile Hypoposphatasia -
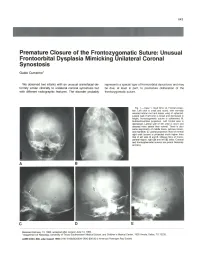
Premature Closure of the Frontozygomatic Suture: Unusual Frontoorbital Dysplasia Mimicking Unilateral Coronal Synostosis
643 Premature Closure of the Frontozygomatic Suture: Unusual Frontoorbital Dysplasia Mimicking Unilateral Coronal Synostosis Guido Currarino 1 We observed two infants with an unusual craniofacial de represents a special type of frontoorbital dysostosis and may formity similar clinically to unilateral coronal synostosis but be due, at least in part, to premature obliteration of the with different radiographic features. The disorder probably frontozygomatic suture. Fig. 1.-Case 1. Skull film s. A, Frontal projec tion. Left orbit is small and round , with normally oriented orbital roof and lesser wing of sphenoid . Lateral wall of left orbit is broad and decreased in height; frontozygomatic suture is obliterated. B, Submentovertical projection. Left frontal area is depressed. Lateral wall of left orbit is short and directed more lateral than normal. There is also some asymmetry of middle fos sa, petrous bones, and mandible. C, Lateral projection. Roof of normal right orbit (arrow) is projected much higher than that of left side. 0 and E, Oblique films of fronto parietal region, right (0) and left (E) sides. Coronal and frontosphenoidal sutures are patent bilaterally (arrows). A B c D E Received February 13, 1984; accepted after revision June 13, 1984. 1 Department of Radiology, University of Texas Southwestern Medical School, and Children's Medical Center, 1935 Amelia, Dallas, TX 75235. AJNR 6:643-646, July/August 1985 0195-6108/85/0604-0643 $00.00 © American Roentgen Ray Society 644 CURRARINO AJNR :6, July/August 1985 A B c Fig . 2.-Case 2. Skull film s. A, Frontal projection. Left orbit is small and pressed. Lateral wall of left orbit is short and directed more laterad than right. -

Tissue-Nonspecific Alkaline Phosphatase
biomolecules Review Tissue-Nonspecific Alkaline Phosphatase— A Gatekeeper of Physiological Conditions in Health and a Modulator of Biological Environments in Disease Daniel Liedtke 1,* , Christine Hofmann 2, Franz Jakob 3, Eva Klopocki 1 and Stephanie Graser 3 1 Institute for Human Genetics, Biocenter, Julius-Maximilians-University Würzburg, 97074 Würzburg, Germany; [email protected] 2 Section of Pediatric Rheumatology and Osteology, University Children’s Hospital of Würzburg, 97080 Würzburg, Germany; [email protected] 3 Bernhard-Heine-Center for Locomotion Research, Julius-Maximilians-University Würzburg, 97076 Würzburg, Germany; [email protected] (F.J.); [email protected] (S.G.) * Correspondence: [email protected] Received: 30 October 2020; Accepted: 5 December 2020; Published: 8 December 2020 Abstract: Tissue-nonspecific alkaline phosphatase (TNAP) is a ubiquitously expressed enzyme that is best known for its role during mineralization processes in bones and skeleton. The enzyme metabolizes phosphate compounds like inorganic pyrophosphate and pyridoxal-50-phosphate to provide, among others, inorganic phosphate for the mineralization and transportable vitamin B6 molecules. Patients with inherited loss of function mutations in the ALPL gene and consequently altered TNAP activity are suffering from the rare metabolic disease hypophosphatasia (HPP). This systemic disease is mainly characterized by impaired bone and dental mineralization but may also be accompanied by neurological symptoms, like anxiety disorders, seizures, and depression. HPP characteristically affects all ages and shows a wide range of clinical symptoms and disease severity, which results in the classification into different clinical subtypes. This review describes the molecular function of TNAP during the mineralization of bones and teeth, further discusses the current knowledge on the enzyme’s role in the nervous system and in sensory perception.