Download ( ~Marcelo/HIV-1 MA Builder, Accessed on 25 July 2021)
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
Final Poster
Associating Finite Groups with Dessins d’Enfants Luis Baeza, Edwin Baeza, Conner Lawrence, and Chenkai Wang Abstract Platonic Solids Rotation Group Dn: Regular Convex Polygon Approach Each finite, connected planar graph has an automorphism group G;such Following Magot and Zvonkin, reduce to easier cases using “hypermaps” permutations can be extended to automorphisms of the Riemann sphere φ : P1(C) P1(C), then composing β = φ f where S 2(R) P1(C). In 1984, Alexander Grothendieck, inspired by a result of f : 1( ) ! 1( )isaBely˘ımapasafunctionofeither◦ zn or ' P C P C Gennadi˘ıBely˘ıfrom 1979, constructed a finite, connected planar graph 4 zn/(zn +1)! 2 such that Aut(f ) Z or Aut(f ) D ,respectively. ' n ' n ∆β via certain rational functions β(z)=p(z)/q(z)bylookingatthe inverse image of the interval from 0 to 1. The automorphisms of such a Hypermaps: Rotation Group Zn graph can be identified with the Galois group Aut(β)oftheassociated 1 1 rational function β : P (C) P (C). In this project, we investigate how Rigid Rotations of the Platonic Solids I Wheel/Pyramids (J1, J2) ! w 3 (w +8) restrictive Grothendieck’s concept of a Dessin d’Enfant is in generating all n 2 I φ(w)= 1 1 z +1 64 (w 1) automorphisms of planar graphs. We discuss the rigid rotations of the We have an action : PSL2(C) P (C) P (C). β(z)= : v = n + n, e =2 n, f =2 − n ◦ ⇥ 2 !n 2 4 zn · Platonic solids (the tetrahedron, cube, octahedron, icosahedron, and I Zn = r r =1 and Dn = r, s s = r =(sr) =1 are the rigid I Cupola (J3, J4, J5) dodecahedron), the Archimedean solids, the Catalan solids, and the rotations of the regular convex polygons,with 4w 4(w 2 20w +105)3 I φ(w)= − ⌦ ↵ ⌦ 1 ↵ Rotation Group A4: Tetrahedron 3 2 Johnson solids via explicit Bely˘ımaps. -

Platonic and Archimedean Geometries in Multicomponent Elastic Membranes
Platonic and Archimedean geometries in multicomponent elastic membranes Graziano Vernizzia,1, Rastko Sknepneka, and Monica Olvera de la Cruza,b,c,2 aDepartment of Materials Science and Engineering, Northwestern University, Evanston, IL 60208; bDepartment of Chemical and Biological Engineering, Northwestern University, Evanston, IL 60208; and cDepartment of Chemistry, Northwestern University, Evanston, IL 60208 Edited by L. Mahadevan, Harvard University, Cambridge, MA, and accepted by the Editorial Board February 8, 2011 (received for review August 30, 2010) Large crystalline molecular shells, such as some viruses and fuller- possible to construct a lattice on the surface of a sphere such enes, buckle spontaneously into icosahedra. Meanwhile multi- that each site has only six neighbors. Consequently, any such crys- component microscopic shells buckle into various polyhedra, as talline lattice will contain defects. If one allows for fivefold observed in many organelles. Although elastic theory explains defects only then, according to the Euler theorem, the minimum one-component icosahedral faceting, the possibility of buckling number of defects is 12 fivefold disclinations. In the absence of into other polyhedra has not been explored. We show here that further defects (21), these 12 disclinations are positioned on the irregular and regular polyhedra, including some Archimedean and vertices of an inscribed icosahedron (22). Because of the presence Platonic polyhedra, arise spontaneously in elastic shells formed of disclinations, the ground state of the shell has a finite strain by more than one component. By formulating a generalized elastic that grows with the shell size. When γ > γà (γà ∼ 154) any flat model for inhomogeneous shells, we demonstrate that coas- fivefold disclination buckles into a conical shape (19). -

The Truncated Icosahedron As an Inflatable Ball
https://doi.org/10.3311/PPar.12375 Creative Commons Attribution b |99 Periodica Polytechnica Architecture, 49(2), pp. 99–108, 2018 The Truncated Icosahedron as an Inflatable Ball Tibor Tarnai1, András Lengyel1* 1 Department of Structural Mechanics, Faculty of Civil Engineering, Budapest University of Technology and Economics, H-1521 Budapest, P.O.B. 91, Hungary * Corresponding author, e-mail: [email protected] Received: 09 April 2018, Accepted: 20 June 2018, Published online: 29 October 2018 Abstract In the late 1930s, an inflatable truncated icosahedral beach-ball was made such that its hexagonal faces were coloured with five different colours. This ball was an unnoticed invention. It appeared more than twenty years earlier than the first truncated icosahedral soccer ball. In connection with the colouring of this beach-ball, the present paper investigates the following problem: How many colourings of the dodecahedron with five colours exist such that all vertices of each face are coloured differently? The paper shows that four ways of colouring exist and refers to other colouring problems, pointing out a defect in the colouring of the original beach-ball. Keywords polyhedron, truncated icosahedron, compound of five tetrahedra, colouring of polyhedra, permutation, inflatable ball 1 Introduction Spherical forms play an important role in different fields – not even among the relics of the Romans who inherited of science and technology, and in different areas of every- many ball games from the Greeks. day life. For example, spherical domes are quite com- The Romans mainly used balls composed of equal mon in architecture, and spherical balls are used in most digonal panels, forming a regular hosohedron (Coxeter, 1973, ball games. -
![Arxiv:1403.3190V4 [Gr-Qc] 18 Jun 2014 ‡ † ∗ Stefloig H Oetintr Ftehmloincon Hamiltonian the of [16]](https://docslib.b-cdn.net/cover/6577/arxiv-1403-3190v4-gr-qc-18-jun-2014-stefloig-h-oetintr-ftehmloincon-hamiltonian-the-of-16-926577.webp)
Arxiv:1403.3190V4 [Gr-Qc] 18 Jun 2014 ‡ † ∗ Stefloig H Oetintr Ftehmloincon Hamiltonian the of [16]
A curvature operator for LQG E. Alesci,∗ M. Assanioussi,† and J. Lewandowski‡ Institute of Theoretical Physics, University of Warsaw (Instytut Fizyki Teoretycznej, Uniwersytet Warszawski), ul. Ho˙za 69, 00-681 Warszawa, Poland, EU We introduce a new operator in Loop Quantum Gravity - the 3D curvature operator - related to the 3-dimensional scalar curvature. The construction is based on Regge Calculus. We define this operator starting from the classical expression of the Regge curvature, we derive its properties and discuss some explicit checks of the semi-classical limit. I. INTRODUCTION Loop Quantum Gravity [1] is a promising candidate to finally realize a quantum description of General Relativity. The theory presents two complementary descriptions based on the canon- ical and the covariant approach (spinfoams) [2]. The first implements the Dirac quantization procedure [3] for GR in Ashtekar-Barbero variables [4] formulated in terms of the so called holonomy-flux algebra [1]: one considers smooth manifolds and defines a system of paths and dual surfaces over which the connection and the electric field can be smeared. The quantiza- tion of the system leads to the full Hilbert space obtained as the projective limit of the Hilbert space defined on a single graph. The second is instead based on the Plebanski formulation [5] of GR, implemented starting from a simplicial decomposition of the manifold, i.e. restricting to piecewise linear flat geometries. Even if the starting point is different (smooth geometry in the first case, piecewise linear in the second) the two formulations share the same kinematics [6] namely the spin-network basis [7] first introduced by Penrose [8]. -

Flippable Tilings of Constant Curvature Surfaces
FLIPPABLE TILINGS OF CONSTANT CURVATURE SURFACES FRANÇOIS FILLASTRE AND JEAN-MARC SCHLENKER Abstract. We call “flippable tilings” of a constant curvature surface a tiling by “black” and “white” faces, so that each edge is adjacent to two black and two white faces (one of each on each side), the black face is forward on the right side and backward on the left side, and it is possible to “flip” the tiling by pushing all black faces forward on the left side and backward on the right side. Among those tilings we distinguish the “symmetric” ones, for which the metric on the surface does not change under the flip. We provide some existence statements, and explain how to parameterize the space of those tilings (with a fixed number of black faces) in different ways. For instance one can glue the white faces only, and obtain a metric with cone singularities which, in the hyperbolic and spherical case, uniquely determines a symmetric tiling. The proofs are based on the geometry of polyhedral surfaces in 3-dimensional spaces modeled either on the sphere or on the anti-de Sitter space. 1. Introduction We are interested here in tilings of a surface which have a striking property: there is a simple specified way to re-arrange the tiles so that a new tiling of a non-singular surface appears. So the objects under study are actually pairs of tilings of a surface, where one tiling is obtained from the other by a simple operation (called a “flip” here) and conversely. The definition is given below. -

Visualization of Regular Maps
Symmetric Tiling of Closed Surfaces: Visualization of Regular Maps Jarke J. van Wijk Dept. of Mathematics and Computer Science Technische Universiteit Eindhoven [email protected] Abstract The first puzzle is trivial: We get a cube, blown up to a sphere, which is an example of a regular map. A cube is 2×4×6 = 48-fold A regular map is a tiling of a closed surface into faces, bounded symmetric, since each triangle shown in Figure 1 can be mapped to by edges that join pairs of vertices, such that these elements exhibit another one by rotation and turning the surface inside-out. We re- a maximal symmetry. For genus 0 and 1 (spheres and tori) it is quire for all solutions that they have such a 2pF -fold symmetry. well known how to generate and present regular maps, the Platonic Puzzle 2 to 4 are not difficult, and lead to other examples: a dodec- solids are a familiar example. We present a method for the gener- ahedron; a beach ball, also known as a hosohedron [Coxeter 1989]; ation of space models of regular maps for genus 2 and higher. The and a torus, obtained by making a 6 × 6 checkerboard, followed by method is based on a generalization of the method for tori. Shapes matching opposite sides. The next puzzles are more challenging. with the proper genus are derived from regular maps by tubifica- tion: edges are replaced by tubes. Tessellations are produced us- ing group theory and hyperbolic geometry. The main results are a generic procedure to produce such tilings, and a collection of in- triguing shapes and images. -
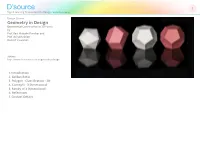
Geometry in Design Geometrical Construction in 3D Forms by Prof
D’source 1 Digital Learning Environment for Design - www.dsource.in Design Course Geometry in Design Geometrical Construction in 3D Forms by Prof. Ravi Mokashi Punekar and Prof. Avinash Shide DoD, IIT Guwahati Source: http://www.dsource.in/course/geometry-design 1. Introduction 2. Golden Ratio 3. Polygon - Classification - 2D 4. Concepts - 3 Dimensional 5. Family of 3 Dimensional 6. References 7. Contact Details D’source 2 Digital Learning Environment for Design - www.dsource.in Design Course Introduction Geometry in Design Geometrical Construction in 3D Forms Geometry is a science that deals with the study of inherent properties of form and space through examining and by understanding relationships of lines, surfaces and solids. These relationships are of several kinds and are seen in Prof. Ravi Mokashi Punekar and forms both natural and man-made. The relationships amongst pure geometric forms possess special properties Prof. Avinash Shide or a certain geometric order by virtue of the inherent configuration of elements that results in various forms DoD, IIT Guwahati of symmetry, proportional systems etc. These configurations have properties that hold irrespective of scale or medium used to express them and can also be arranged in a hierarchy from the totally regular to the amorphous where formal characteristics are lost. The objectives of this course are to study these inherent properties of form and space through understanding relationships of lines, surfaces and solids. This course will enable understanding basic geometric relationships, Source: both 2D and 3D, through a process of exploration and analysis. Concepts are supported with 3Dim visualization http://www.dsource.in/course/geometry-design/in- of models to understand the construction of the family of geometric forms and space interrelationships. -

Visualization of Regular Maps Scientific Visualization – Math Vis Vis – Human Computer Interaction – Visual Analytics – Parallel
2/2/2016 Visualization Eindhoven – information visualization – software visualization – perception – geographic visualization Visualization of Regular Maps scientific visualization – math vis vis – human computer interaction – visual analytics – parallel Jarke J. van Wijk Eindhoven University of Technology JCB 2016, Bordeaux Data : flow fields – trees – graphs – tables – mobile data – events – … Applications : software analysis – business cases – bioinformatics – … Can you draw a Seifert surface? Eindhoven 2004 Huh? Starting MathVis Arjeh Cohen Me Discrete geometry, Visualization algebra Seifert surface Eindhoven 2006 1 2/2/2016 Can you show 364 triangles in 3D? Aberdeenshire Sure! 2000 BC I’ll give you the Arjeh Cohenpattern, you can Me pick a nice shape. Discrete geometry, Visualization algebra Ok! Athens 450 BC Scottish Neolithic carved stone balls 8 12 20 8 12 20? 6 6 4 4 Platonic solids: perfectly symmetric Platonic solids: perfectly symmetric 2 2/2/2016 How to get more faces, How to get more faces, all perfectly symmetric? all perfectly symmetric? Use shapes with holes Aim only at topological symmetry 64 64 #faces? Königsberg 1893 3 2/2/2016 Adolf Hurwitz 1859-1919 max. #faces: 3 28(genus – 1) 7 Genus Faces Genus Faces 3 56 3 56 7 168 7 168 14 364 14 364 … … … … Can you show 364 triangles in 3D? New Orleans Sure! 2009 I’ll give you the three years later Arjeh pattern, you can Me Cohen pick a nice shape. Ok! 4 2/2/2016 The general puzzle Construct space models of regular maps Symmetric Tiling of Closed Surfaces: Visualization of -

Spherical Trihedral Metallo-Borospherenes
Lawrence Berkeley National Laboratory Recent Work Title Spherical trihedral metallo-borospherenes. Permalink https://escholarship.org/uc/item/59z4w76k Journal Nature communications, 11(1) ISSN 2041-1723 Authors Chen, Teng-Teng Li, Wan-Lu Chen, Wei-Jia et al. Publication Date 2020-06-02 DOI 10.1038/s41467-020-16532-x Peer reviewed eScholarship.org Powered by the California Digital Library University of California ARTICLE https://doi.org/10.1038/s41467-020-16532-x OPEN Spherical trihedral metallo-borospherenes ✉ ✉ Teng-Teng Chen 1,5, Wan-Lu Li2,5 , Wei-Jia Chen1, Xiao-Hu Yu 3, Xin-Ran Dong4, Jun Li 2,4 & ✉ Lai-Sheng Wang 1 The discovery of borospherenes unveiled the capacity of boron to form fullerene-like cage structures. While fullerenes are known to entrap metal atoms to form endohedral metallo- fullerenes, few metal atoms have been observed to be part of the fullerene cages. Here we report the observation of a class of remarkable metallo-borospherenes, where metal atoms 1234567890():,; – – are integral parts of the cage surface. We have produced La3B18 and Tb3B18 and probed their structures and bonding using photoelectron spectroscopy and theoretical calculations. – Global minimum searches revealed that the most stable structures of Ln3B18 are hollow – cages with D3h symmetry. The B18-framework in the Ln3B18 cages can be viewed as con- sisting of two triangular B6 motifs connected by three B2 units, forming three shared B10 rings which are coordinated to the three Ln atoms on the cage surface. These metallo- borospherenes represent a new class of unusual geometry that has not been observed in chemistry heretofore. -

Regular Cyclic Coverings of the Platonic Maps* GARETH A. JONES
Regular Cyclic Coverings of the Platonic Maps* GARETH A. JONES and DAVID B. SUROWSKI** We use homological methods to describe the regular maps and hypermaps which are cyclic coverings of the Platonic maps, branched over the face-centers, vertices or midpoints of edges. 1. INTRODUCTION The M¨obius-Kantor map {4 + 4, 3} [CMo, §8.8, 8.9] is a regular orientable map of type {8, 3} and genus 2. It is a 2-sheeted covering of the cube {4, 3}, branched over the centers of its six faces, each of which lifts to an octagonal face. Its (orientation-preserving) automorphism ∼ group is isomorphic to GL2(3), a double covering of the automorphism group P GL2(3) = S4 of the cube. The aim of this note is to describe all the regular maps and hypermaps which can be obtained in a similar manner as cyclic branched coverings of the Platonic maps M, with the branching at the face-centers, vertices, or midpoints of edges. The method used is to consider the action of Aut M on certain homology modules; in a companion paper [SJ] we use cohomological techniques to give explicit constructions of these coverings in terms of voltage assignments. Let M be a Platonic map, that is, a regular map on the sphere S2. In the notation of [CMo], M has type {n, m}, or simply M = {n, m}, where the faces are n-gons and the vertices have valency m; as a hypermap, M has type (m, 2, n). Here 0 ≤ (m − 2)(n − 2) < 4, so M is the dihedron {n, 2}, the hosohedron {2, m}, the tetrahedron {3, 3}, the cube {4, 3}, the octahedron {3, 4}, the dodecahedron {5, 3} or the icosahedron {3, 5}. -

Nn636jn0614.Pdf
/ (41) HPP-72-5 // OrPRINTS Genetics 194 / BIOCHEMICAL' pECTPOMEW " BY R. WAIUH JOHN WILEY 1372 CHAPTER 7 USE OF A COMPUTER TO IDENTIFY UNKNOWN COMPOUNDS: THE AUTO- MATION OF SCIENTIFIC INFERENCE* JOSHUA LEDERBERG Departmentof Genetics, School of Medicine, Stanford University, Stanford, California Introduction B. Motivation 194 C. Implementation 194 1. Generator 194 2. All the Ways to Build a Molecule , 195 3. Graphs of Ring Compounds 197 4. Heuristics 197 D. Commentary 199 E. Example 200 A. INTRODUCTION knowledge. The problem was merely one of selecting the proper texts. The Argentinian writer JorgeLuis Borges.in a short The identification of an unknown compound story called "The Library of Babel," showed that all presents a similar challenge. If the universe of knowledge can be reduced to a problem of selection. possibilities were the problem might not be He portrayed a library of infinite dimensions filled rigorously soluble. Practical solutions depend upon with books printed in an obscure code in which the ingenuity with which the domain of acceptable familiar phrases occasionally appeared. Eventually, solutions can be narrowed within a particular experi- a mathematician-inhabitant of this space surmised mental context and the efficiency with which tentative that each book was one of all possible random con- solutions can be tested against the data. catenations of letters. After a few centuries of dis- The previous chapter deals with the pragmatics couragement, the inhabitants were inspired by a new of searching the index to a finite library, i.e., the revelation — that the library must in fact contain all catalog of mass spectra of previously studied mole- cules, with occasional extensions to related structures. -

Title of the Article
Symmetry: Culture and Science Vol. x, No.x, page_first-page_last, 2013 SYMMETRICAL IMMERSIONS OF LOW-GENUS NON-ORIENTABLE REGULAR MAPS Carlo H. Séquin Computer Scientist, (b. Winterthur, Switzerland, 1941). Address: EECS Computer Science, U.C. Berkeley, CA. 94720, U.S.A. E-mail: [email protected] Fields of interest: Computer Graphics, Computer-Aided Design, Math Visualization, Artistic Geometry. Awards: IEEE Technical Achievement Award, 2003; McEntyre Award for Excellence in Teaching, 1996. Publications and/or Exhibitions: *C. H. Séquin, "Symmetrical Hamiltonian Manifolds on Regular 3D and 4d Polytopes" Coxeter Day, Banff, Canada, Aug.3, 2005, pp 463-472. C. H. Séquin, "Hilbert Cube 512," Artist's Sketch, SIGGRAPH'06, Boston, July 30 - Aug. 3, 2006. C. H. Séquin, "Patterns on the Genus-3 Klein Quartic," Proc. BRIDGES Conference, London, Aug. 4-9, 2006, pp 245-254. C. H. Séquin and Jaron Lanier, "Hyper-Seeing the Regular Hendeca-choron," ISAMA Proc. pp159-166, Texas A&M, May 17-21, 2007. *C. H. Séquin, "Symmetric Embedding of Locally Regular Hyperbolic Tilings," Bridges Conference, San Sebastian, Spain, July 24-27, 2007. C. H. Séquin and J. F. Hamlin, "The Regular 4-Dimensional 57-Cell," SIGGRAPH'07, Sketches and Applications, San Diego, Aug. 4-9, 2007. C. H. Séquin, "Eightfold Way," Gathering for Gardner G4G8, Atlanta GA, March 27-30, 2008 *C. H. Séquin, "Intricate Isohedral Tilings of 3D Euclidean Space," Bridges Conference, Leeuwarden, The Netherlands, July 24-28, 2008, pp 139-148. *M. Howison and C. H. Séquin, "CAD Tools for the Construction of 3D Escher Tiles," Computer-Aided Design and Applications, Vol 6, No 6, pp 737-748, 2009.