Novel Structure-Based Profiling and Annotation Workflow—High-Throughput Analysis of Flavonoids Using the Thermo Scientific Orbitrap ID-X Tribrid MS
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Dedicated to the Memory of Ludovico Sorrentino, Emeritus Pharmacology Professor (1932-2010)
27/2/2020 PharmacologyOnLine | Front Dedicated to the Memory of Ludovico Sorrentino, Emeritus Pharmacology Professor (1932-2010) ISSN: 1827-8620 Fri, 28 Feb 2020 03:27:58 +0100 PhOL - PharmacologyOnLine Pharmacologyonline is an international journal online. It is interdisciplinary and aims to bring together different approaches for a diverse readership all over the world in Pharmacology, Ethnopharmacology and Medicinal Plants. High quality papers will be accepted dealing with any aspect of Pharmacology. Pharmacologyonline will provide a medium for the publication of high quality, peer-reviewed, original research articles, reviews and case reports in the field of Pharmacology, Ethnopharmacology and Medicinal Plants. It will be published quarterly. Pharmacologyonline accepts submissions, presented as original research (Archives and Young Researchers section), short papers, reviews, correspondence (Newsletter section) and case reports (Case Report section). Submissions regarding all types of papers are welcomed for publication. In general, all articles submitted are peer reviewed by at least two researchers expert in the field. Pharmacologyonline is an “Open Access Journal”! All articles published are made freely and permanently accessible online immediately upon publication, without subscription charges or registration barriers. Authors of articles published in Pharmacologyonline are the copyright holders of their articles and have granted to any third party, in advance and in perpetuity, the right to use, reproduce or disseminate the article, according to the SILAE copyright and license agreement. Article-processing charges Open access publishing is not without costs. Pharmacologyonline therefore levies an article-processing charge of €300 (Euro) / $387 (USD) for authors from European Union, U.S.A., Japan and €120 (Euro) / $155 (USD) for others countries for each article accepted for publication. -

Molecular Mechanisms of the Anti-Obesity and Anti-Diabetic Properties of Flavonoids
International Journal of Molecular Sciences Review Molecular Mechanisms of the Anti-Obesity and Anti-Diabetic Properties of Flavonoids Mohammed Kawser Hossain, Ahmed Abdal Dayem, Jihae Han, Yingfu Yin, Kyeongseok Kim, Subbroto Kumar Saha, Gwang-Mo Yang, Hye Yeon Choi and Ssang-Goo Cho * Department of Animal Biotechnology, Animal Resources Research Center, Incurable Disease Animal Model and Stem Cell Institute (IDASI), Konkuk University, Gwangjin-gu, Seoul 05029, Korea; [email protected] (M.K.H.); [email protected] (A.A.D.); [email protected] (J.H.); [email protected] (Y.Y.); [email protected] (K.K.); [email protected] (S.K.S.); [email protected] (G.-M.Y.); [email protected] (H.Y.C.) * Correspondence: [email protected]; Tel.: +82-2-450-4207 Academic Editor: Toshiro Arai Received: 12 March 2016; Accepted: 6 April 2016; Published: 15 April 2016 Abstract: Obesity and diabetes are the most prevailing health concerns worldwide and their incidence is increasing at a high rate, resulting in enormous social costs. Obesity is a complex disease commonly accompanied by insulin resistance and increases in oxidative stress and inflammatory marker expression, leading to augmented fat mass in the body. Diabetes mellitus (DM) is a metabolic disorder characterized by the destruction of pancreatic β cells or diminished insulin secretion and action insulin. Obesity causes the development of metabolic disorders such as DM, hypertension, cardiovascular diseases, and inflammation-based pathologies. Flavonoids are the secondary metabolites of plants and have 15-carbon skeleton structures containing two phenyl rings and a heterocyclic ring. More than 5000 naturally occurring flavonoids have been reported from various plants and have been found to possess many beneficial effects with advantages over chemical treatments. -

Variability of Hordeum Vulgare L. Cultivars in Yield, Antioxidant Potential, and Cholinesterase Inhibitory Activity
sustainability Article Variability of Hordeum vulgare L. Cultivars in Yield, Antioxidant Potential, and Cholinesterase Inhibitory Activity Joanna Kobus-Cisowska 1, Piotr Szulc 2,*, Oskar Szczepaniak 1 , Marcin Dziedzi ´nski 1 , Daria Szymanowska 3 , Krystyna Szymandera-Buszka 1 , El˙zbietaGory ´nska-Goldmann 4 , Michał Gazdecki 4, Aleksandra Telichowska 1 and Marta Ligaj 5 1 Department of Gastronomy Sciences and Functional Foods, Pozna´nUniversity of Life Sciences, 60-637 Pozna´n,Poland; [email protected] (J.K.-C.); [email protected] (O.S.); [email protected] (M.D.); [email protected] (K.S.-B.); [email protected] (A.T.) 2 Department of Agronomy, Pozna´nUniversity of Life Sciences, 60-637 Pozna´n,Poland 3 Department of Biotechnology and Food Microbiology, Pozna´nUniversity of Life Sciences, 60-637 Pozna´n, Poland; [email protected] 4 Department of Economics and Economy Policy in Agribusiness, Pozna´nUniversity of Life Sciences, 60-637 Pozna´n,Poland; [email protected] (E.G.-G.); [email protected] (M.G.) 5 Department of Industrial Products and Packaging, Pozna´nUniversity of Economics and Business, 61-875 Pozna´n,Poland; [email protected] * Correspondence: [email protected]; Tel.: +48-61-848-7515 Received: 8 February 2020; Accepted: 29 February 2020; Published: 3 March 2020 Abstract: Barley (Hordeum vulgare L.) is an annual plant cultivated in spring or autumn. Currently, over 70% of the cultivated barley grains are utilized for preparing fodder, while the rest is used for the production of malt and cereals in the food industry. -

Quercetin Gregory S
amr Monograph Quercetin Gregory S. Kelly, ND Description and Chemical Composition 5, 7, 3’, and 4’ (Figure 2). The difference between quercetin and kaempferol is that the latter lacks Quercetin is categorized as a flavonol, one of the the OH group at position 3’. The difference six subclasses of flavonoid compounds (Table 1). between quercetin and myricetin is that the latter Flavonoids are a family of plant compounds that has an extra OH group at position 5’. share a similar flavone backbone (a three-ringed By definition quercetin is an aglycone, lacking an molecule with hydroxyl [OH] groups attached). A attached sugar. It is a brilliant citron yellow color multitude of other substitutions can occur, giving and is entirely insoluble in cold water, poorly rise to the subclasses of flavonoids and the soluble in hot water, but quite soluble in alcohol different compounds found within these subclasses. Flavonoids also occur as either glycosides (with attached sugars [glycosyl groups]) or as aglycones (without attached sugars).1 Figure 1. 3-Hydroxyflavone Backbone with Flavonols are present in a wide variety of fruits Locations Numbered for Possible Attachment and vegetables. In Western populations, estimated of Hydroxyl (OH) and Glycosyl Groups daily intake of flavonols is in the range of 20-50 mg/day.2 Most of the dietary intake is as 3’ flavonol glycosides of quercetin, kaempferol, and 2’ 4’ myricetin rather than their aglycone forms (Table 8 1 2). Of this, about 13.82 mg/day is in the form of O 1’ 2 7 5’ quercetin-type flavonols. 2 The variety of dietary flavonols is created by the 6’ 6 differential placement of phenolic-OH groups and 3 OH attached sugars. -

WO 2018/002916 Al O
(12) INTERNATIONAL APPLICATION PUBLISHED UNDER THE PATENT COOPERATION TREATY (PCT) (19) World Intellectual Property Organization International Bureau (10) International Publication Number (43) International Publication Date WO 2018/002916 Al 04 January 2018 (04.01.2018) W !P O PCT (51) International Patent Classification: (81) Designated States (unless otherwise indicated, for every C08F2/32 (2006.01) C08J 9/00 (2006.01) kind of national protection available): AE, AG, AL, AM, C08G 18/08 (2006.01) AO, AT, AU, AZ, BA, BB, BG, BH, BN, BR, BW, BY, BZ, CA, CH, CL, CN, CO, CR, CU, CZ, DE, DJ, DK, DM, DO, (21) International Application Number: DZ, EC, EE, EG, ES, FI, GB, GD, GE, GH, GM, GT, HN, PCT/IL20 17/050706 HR, HU, ID, IL, IN, IR, IS, JO, JP, KE, KG, KH, KN, KP, (22) International Filing Date: KR, KW, KZ, LA, LC, LK, LR, LS, LU, LY, MA, MD, ME, 26 June 2017 (26.06.2017) MG, MK, MN, MW, MX, MY, MZ, NA, NG, NI, NO, NZ, OM, PA, PE, PG, PH, PL, PT, QA, RO, RS, RU, RW, SA, (25) Filing Language: English SC, SD, SE, SG, SK, SL, SM, ST, SV, SY, TH, TJ, TM, TN, (26) Publication Language: English TR, TT, TZ, UA, UG, US, UZ, VC, VN, ZA, ZM, ZW. (30) Priority Data: (84) Designated States (unless otherwise indicated, for every 246468 26 June 2016 (26.06.2016) IL kind of regional protection available): ARIPO (BW, GH, GM, KE, LR, LS, MW, MZ, NA, RW, SD, SL, ST, SZ, TZ, (71) Applicant: TECHNION RESEARCH & DEVEL¬ UG, ZM, ZW), Eurasian (AM, AZ, BY, KG, KZ, RU, TJ, OPMENT FOUNDATION LIMITED [IL/IL]; Senate TM), European (AL, AT, BE, BG, CH, CY, CZ, DE, DK, House, Technion City, 3200004 Haifa (IL). -

Flavone and Flavonol Glycosides from the Leaves of Triumfetta Procumbens in Ryukyu Islands
Bull. Natl. Mus. Nat. Sci., Ser. B, 38(2), pp. 63–67, May 22, 2012 Flavone and Flavonol Glycosides from the Leaves of Triumfetta procumbens in Ryukyu Islands Tsukasa Iwashina* and Goro Kokubugata Department of Botany, National Museum of Nature and Science, 4–1–1 Amakubo, Tsukuba, Ibaraki 305–0005, Japan * E-mail: [email protected] (Received 11 February 2012; accepted 28 March 2012) Abstract Four flavonoids were isolated from the leaves of Triumfetta procumbens in Ryukyu Islands for the first time. They were identified as apigenin 7-O-glucuronide, luteolin 7-O-glucuro- nide, schaftoside and kaempferol 3-O-(p-coumaroylglucoside) by UV, LC-MS, acid hydrolysis, and direct TLC and HPLC comparisons with authentic samples. From the species belonging to the family Tiliaceae sensu stricto including the genus Triumfetta, some flavonols and flavones have been found together with a few dihydroflavonols and proanthocyanidins. However, three flavonoid O-glycosides and C-glycoside except for kaempferol 3-O-(p-coumaroylglucoside), which were isolated in this experiment, were reported from the family Tiliaceae s.s. for the first time. Key words : apigenin and luteolin 7-glucuronides, flavonoids, kaempferol 3-(p-coumaroylgluco- side), schaftoside, Tiliaceae s.s., Triumfetta. 7-O-rhamnosylarabinoside and scutellarein Introduction 7-O-rhamnoside, respectively (Srinivasan and The genus Triumfetta belongs to the family Subramanian, 1981), chemical structure of trium- Tiliaceae, which was recently included in the boidin was later revised as scutellarein 6-O-β-D- family Malvaceae together with Bombacaceae xyloside-7-O-α-L-rhamnopyranoside by 1H and and Sterculiaceae by APG. The genus consists of 13C NMR and FAB-MS methods (Nair et al., ca. -
Reference Substances 2018/2019
Reference Substances 2018 / 2019 Reference Substances Reference 2018/2019 Contents | 3 Contents Page Welcome 4 Our Services 5 Reference Substances 6 Index I: Alphabetical List of Reference Substances and Synonyms 156 Index II: Plant-specific Marker Compounds 176 Index III: CAS Registry Numbers 214 Index IV: Substance Classification 224 Our Reference Substance Team 234 Order Information 237 Order Form 238 Prices insert 4 | Welcome Welcome to our new 2018 / 2019 catalogue! PhytoLab proudly presents the new you will also be able to view exemplary Index I contains an alphabetical list of all 2018 / 2019 catalogue of phyproof® certificates of analysis and download substances and their synonyms. It pro- Reference Substances. The seventh edition material safety data sheets (MSDS). vides information which name of a refer- of our catalogue now contains well over ence substance is used in this catalogue 1300 phytochemicals. As part of our We very much hope that our product and guides you directly to the correct mission to be your leading supplier of portfolio meets your expectations. The list page. herbal reference substances PhytoLab of substances will be expanded even has characterized them as primary further in the future, based upon current If you are a planning to analyse a specific reference substances and will supply regulatory requirements and new scientific plant please look for the botanical them together with the comprehensive developments. The most recent information name in Index II. It will inform you about certificates of analysis you are familiar will always be available on our web site. common marker compounds for this herb. with. -

Extraction of Kaempferol and Its Glycosides Using Supercritical Fluids from Plant Sources: a Review
review ISSN 1330-9862 doi: 10.17113/ftb.56.04.18.5870 Extraction of Kaempferol and Its Glycosides Using Supercritical Fluids from Plant Sources: A Review Sandro Cid-Ortega1* SUMMARY and José Alberto Research has focused on the study and design of new products containing phyto- Monroy-Rivera2† chemicals (polyphenolic acids, flavonoids and anthocyanins, among others) that contrib- ute to a better life quality for consumers, as well as techniques to improve the extraction of these compounds. In this regard, extraction with supercritical fluids (usually CO2) is a 1 Technological University of Izúcar technology that has better advantages than organic solvents, since lower temperatures de Matamoros (UTIM), Department that are used do not damage the product, and is more effective at high pressures. Extracts of Food Process Engineering, from plant sources contain significant concentrations of flavonols, one of the main com- Prolongación Reforma No. 168, Barrio pounds being kaempferol. Research has proven the antioxidant, anticarcinogenic, antidi- de Santiago Mihuacán 74420, Izúcar de Matamoros, Puebla, Mexico abetic, cardioprotective, neuroprotective and antimicrobial properties of kaempferol and 2 Technological National of Mexico/ its glycosides. This review aims to present the main functional properties and extraction Technological Institute of Veracruz, with supercritical fluids of kaempferol and its glycosides, as well as the application of this Research and Development Unit in technology in the food area. Foods (UNIDA), Calzada Miguel Ángel de Quevedo No. 2779, Formando Hogar 91897, Veracruz, Veracruz, Key words: phytochemicals, flavonols, kaempferol, supercritical fluids Mexico INTRODUCTION Received: 28 May 2018 The use of medicinal plants in Mexico is still important in traditional medicine for the Accepted: 4 October 2018 treatment of various diseases among the population and these plants are also an import- ant source for development of new medicines (1–3). -

Korean Journal of Environmental Agriculture
Korean Journal of Environmental Agriculture Korean J Environ Agric. 2017;36(4):249-255. English Online ISSN: 2233-4173 Published online 2017 December 6. https://doi.org/10.5338/KJEA.2017.36.4.39 Print ISSN: 1225-3537 Research Article Open Access Quali-Quantitative Analysis of Flavonoids for Mulberry Leaf and Fruit of‘Suhyang’ Wan-Taek Ju1,O-ChulKwon1,Min-KiLee2,Hyun-BokKim1, Gyoo-Byung Sung1 and Yong-Soon Kim1* 1Department of Agricultural Biology, Sericulture & Apiculture Division, National Institute of Agricultural Science, Rural Development Administration, Wanju, 55365, Korea 2Department of Agro-food Resources, Functional Food and Nutrition Division, National Institute of Agricultural Science, Rural Development Administration, Wanju, 55365, Korea Received: 6 November 2017 / Revised: 7 November 2017 / Accepted: 28 November 2017 ORCID Copyrightⓒ 2017 The Korean Society of Environmental Agriculture Wan-Taek Ju This is an Open-Access article distributed under the terms of the Creative Commons Attribution http://orcid.org/0000-0002-0752-470X Non-Commercial License (http://creativecommons.org/licenses/by-nc/3.0) which permits unrestricted Yong-Soon Kim non-commercial use, distribution, and reproduction in any medium, provided the original work is http://orcid.org/0000-0003-0799-9278 properly cited. Abstract analyzed from Suhyang using UPLC-DAD-QTOF/MS chromatogram. To the best of our knowledge, Quercetin BACKGROUND: Globally, mulberry (Morus sp.) is 3-O-(6''-O-malonyl) glucoside and quercetin 3-O-rutinoside exploited for feeding leaf to silkworms in order to obtain (rutin) was detected on the highest content in leaf and fruit, silk fiber or for animal feedstock production. Also, respectively and further research will be devoted to evaluate mulberry fruit is known to a by-product that was produced their biological activity. -
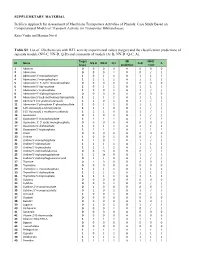
SUPPLEMETARY MATERIAL in Silico Approach for Assessment Of
SUPPLEMETARY MATERIAL In Silico Approach for Assessment of Membrane Transporters Activities of Phenols: Case Study Based on Computational Models of Transport Activity for Transporter Bilitranslocase Katja Venko and Marjana Novič Table S1: List of 120 chemicals with BTL activity experimental values (target) and the classification predictions of separate models (NN-C, NN-D, Q-D) and consensus of models (A+B, NN-D+Q-C, A). Target AB NN-D ID Name NN-D NN-D Q-C A+B A (exp.) prediction + Q-D 1 Adenine 0 0 0 0 A 0 0 0 2 Adenosine 0 0 0 1 B 0 / / 3 Adenosine 3’-monophoshate 1 0 1 1 B 1 1 / 4 Adenosine 5’-monophoshate 1 1 1 1 A 1 1 1 5 Adenosine 3′, 5′-cyclic monophosphate 0 0 0 0 A 0 0 0 6 Adenosine 5′-diphosphate 1 0 1 1 B 1 1 / 7 Adenosine 5′-triphosphate 1 1 0 1 B 1 / / 8 Adenosine-5′-diphosphoglucose 0 0 0 0 A 0 0 0 9 Adenosine 5'-(α,β-methylene) diphosphate 1 1 1 1 A 1 1 1 10 Adenine 9-β-D-arabinofuranoside 1 1 0 1 B 1 / / 11 Adenosine 3′-phosphate 5′-phosphosulfate 1 0 1 1 B 1 1 / 12 S-(5′-Adenosyl)-L-homocysteine 1 1 1 0 B 1 / / 13 S-(5′-Adenosyl)-L-methionine chloride 1 1 1 0 B 1 / / 14 Guanosine 0 1 0 1 B 1 / / 15 Guanosine 5′-monophosphate 1 1 1 1 A 1 1 1 16 Guanosine 3′. -

Reference Substances
Reference Substances 2020/2021 Contents | 3 Contents Page Welcome 4 Our Services 5 Reference Substances 6 Index I: Alphabetical List of Reference Substances and Synonyms 168 Index II: CAS Registry Numbers 190 Index III: Substance Classification 200 Our Reference Substance Team 212 Distributors & Area Representatives 213 Ordering Information 216 Order Form 226 4 | Welcome Welcome to our new 2020 / 2021 catalogue! PhytoLab proudly presents the new for all reference substances are available Index I contains an alphabetical list of 2020/2021 catalogue of phyproof® for download. all substances and their synonyms. It Reference Substances. The eighth edition provides information which name of a of our catalogue now contains well over We very much hope that our product reference substance is used in this 1400 natural products. As part of our portfolio meets your expectations. The catalogue and guides you directly to mission to be your leading supplier of list of substances will be expanded even the correct page. herbal reference substances PhytoLab further in the future, based upon current has characterized them as primary regulatory requirements and new Index II contains a list of the CAS registry reference substances and will supply scientific developments. The most recent numbers for each reference substance. them together with the comprehensive information will always be available on certificates of analysis you are familiar our web site. However, if our product list Finally, in Index III we have sorted all with. does not include the substance you are reference substances by structure based looking for please do not hesitate to get on the class of natural compounds that Our phyproof® Reference Substances will in touch with us. -

Flavonoids from Theligonum Japonicum Endemic to Japan and Their Chemical Character
Bull. Natl. Mus. Nat. Sci., Ser. B, 43(1), pp. 27–32, February 22, 2017 Flavonoids from Theligonum japonicum Endemic to Japan and their Chemical Character Tsukasa Iwashina Department of Botany, National Museum of Nature and Science, Amakubo 4–1–1, Tsukuba, Ibaraki 305–0005, Japan E-mail: [email protected] (Received 20 November 2016; accepted 21 December 2016) Abstract Flavonoids from the aerial parts of Theligonum japonicum endemic to Japan were sur- veyed. Eleven flavonoids were found and isolated by various chromatography including paper, column and HPLC. Of their compounds, four were identified as quercetin 3-O-glucoside, quercetin 3-O-galactoside, isorhamnetin 3-O-glucoside and isorhamnetin 3-O-galactoside by UV, LC-MS, acid hydrolysis and TLC and HPLC comparisons with authentic samples. Other seven were char- acterized as 3-O-hexosides and 3-O-rhamnosylhexosides of quercetin and isorhamnetin which attached 1 mol unknown substance of molecular weight 162. Although the genus belonged to the family Theligonaceae, it was recently incorporated to the family Rubiaceae by APG III. In this paper, flavonoid character of T. japonicum was compared with Rubia and Hedyotis species which phylogenetically or morphologically related to Theligonum. Key words: chemosystematics, flavonoids, flavonols, Rubiaceae, Theligonaceae, Theligonum japonicum. ligonum species was never reported. As a series Introduction of flavonoid survey of Japanese endemic and The genus Theligonum consists of four spe- endangered plants, in this paper, the isolation and cies, i.e. T. cynocrambe L. in the Mediterranean characterization of flavonoid and related com- and Macronesia, T. macranthum Franch. in pounds from Theligonum japonicum are reported China, T.