Genetic Diversity and Chromosome Complement of Galictis Cuja
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

APROXIMACIÓN AL LENGUAJE REPERTORIAL DE LAS ESTEREOTIPIAS EN MAMIFEROS CARNIVOROS JAGUAR (Panthera Onca), GRISÓN (Galictis
APROXIMACIÓN AL LENGUAJE REPERTORIAL DE LAS ESTEREOTIPIAS EN MAMIFEROS CARNIVOROS JAGUAR (Panthera onca), GRISÓN (Galictis vittata) Y OCELOTE (Leopardus pardalis) DEL ZOOLOGICO GUATIKA, BOYACA – TIBASOSA. Laura Inés Monsalve Zuleta CÓDIGO:2014101101 Trabajo de investigación presentado como requisito parcial para optar al título de: Médica veterinaria especialista en Medicina Homeopática veterinaria Director (a): Néstor Alberto Calderón Maldonado Médico Veterinario – Universidad de la Salle Certificado en Medicina Veterinaria Homeopática – FICH “Luis G. Páez” Especialista en Bioética – Universidad El Bosque Fundación Universitaria Escuela Colombiana de Medicina Homeopática “Luis G. Páez”. Programa de Medicina Homeopática Veterinaria Bogotá, Colombia 2016 2 DEDICATORIA A mi familia, amigos y doctores que me apoyaron en esta etapa de mi vida tanto profesional como personal. 3 AGRADECIMIENTOS Agradezco a mi familia, amigos y Doctores que me ayudaron a culminar esta etapa de mi vida. 4 TABLA DE CONTENIDO: 1. RESUMEN ...................................................................................................................................... 7 2. INTRODUCCIÓN ........................................................................................................................... 8 3. JUSTIFICACIÓN ........................................................................................................................... 9 4. ESTADO DEL ARTE ................................................................................................................. -

Predicting Greater Grison Galictis Vittata Presence from Scarce Records in the Department of Cordoba, Colombia
ORIGINAL CONTRIBUTION Predicting Greater Grison Galictis vittata presence from scarce records in the department of Cordoba, Colombia José F. GONZÁLEZ-MAYA1*, Julio J. CHACÓN PACHECO2, Javier RACERO-CASARRUBIA2, Erika HUMANEZ-LÓPEZ2 & Andrés ARIAS-ALZATE3 1. Proyecto de Conservación de Aguas y Tierras, ProCAT Colombia/Internacional, Calle 97ª # 10-62, Of. 202, Bogotá, Abstract. Colombia. The Greater Grison, Galictis vittata, is a poorly known species in Colombia. Throughout 2. Grupo de Investigación its range major knowledge gaps exist regarding its ecology and conservation. To compile Biodiversidad Unicórdoba, and analyse information about the species´ distribution records in the department of Universidad de Córdoba, Cordoba, Colombia and assess its presence probability according to landscape attributes, Montería, Córdoba, Colombia. we conducted a literature review of all wildlife studies in the region and compiled all possible direct presence records of the species in the department. We generated random 3.Grupo de Mastozoología, location points and characterized each distribution and random location by their distance to landscape attributes and land-cover type and modelled landscape presence using a Instituto de Biología, Multiple Logistic Regression approach. We found 33 records of the species in Cordoba Universidad de Antioquía. with most of the records distributed in the subregion of Alto Sinú (36%). Higher presence Medellín, Colombia. probabilities are localized in areas near forests mostly in the southern parts of the department, showing the species is still related with the largest forest blocks. Grisons Correspondence: appears to potentially tolerate some levels of disturbance but is still dependent to forest. The influence of natural habitats and abundance across the department and other areas José F. -

Controlled Animals
Environment and Sustainable Resource Development Fish and Wildlife Policy Division Controlled Animals Wildlife Regulation, Schedule 5, Part 1-4: Controlled Animals Subject to the Wildlife Act, a person must not be in possession of a wildlife or controlled animal unless authorized by a permit to do so, the animal was lawfully acquired, was lawfully exported from a jurisdiction outside of Alberta and was lawfully imported into Alberta. NOTES: 1 Animals listed in this Schedule, as a general rule, are described in the left hand column by reference to common or descriptive names and in the right hand column by reference to scientific names. But, in the event of any conflict as to the kind of animals that are listed, a scientific name in the right hand column prevails over the corresponding common or descriptive name in the left hand column. 2 Also included in this Schedule is any animal that is the hybrid offspring resulting from the crossing, whether before or after the commencement of this Schedule, of 2 animals at least one of which is or was an animal of a kind that is a controlled animal by virtue of this Schedule. 3 This Schedule excludes all wildlife animals, and therefore if a wildlife animal would, but for this Note, be included in this Schedule, it is hereby excluded from being a controlled animal. Part 1 Mammals (Class Mammalia) 1. AMERICAN OPOSSUMS (Family Didelphidae) Virginia Opossum Didelphis virginiana 2. SHREWS (Family Soricidae) Long-tailed Shrews Genus Sorex Arboreal Brown-toothed Shrew Episoriculus macrurus North American Least Shrew Cryptotis parva Old World Water Shrews Genus Neomys Ussuri White-toothed Shrew Crocidura lasiura Greater White-toothed Shrew Crocidura russula Siberian Shrew Crocidura sibirica Piebald Shrew Diplomesodon pulchellum 3. -

Galictis Cuja Molina, 1782) As Host of Dioctophyme Renale Goeze, 1782 Furão Pequeno (Galictis Cuja Molina, 1782) Como Hospedeiro De Dioctophyme Renale Goeze, 1782
ANIMAL PARASITOLOGY / SCIENTIFIC COMMUNICATION DOI: 10.1590/1808-1657000312016 Lesser Grison (Galictis cuja Molina, 1782) as host of Dioctophyme renale Goeze, 1782 Furão Pequeno (Galictis cuja Molina, 1782) como hospedeiro de Dioctophyme renale Goeze, 1782 Daniela Pedrassani1*, Mayana Worm1, Jéssica Drechmer1, Margareth Cristina Iazzetti Santos1 ABSTRACT: The Dioctophyme renale is a helminth parasite RESUMO: O Dioctophyme renale é um helminto parasita renal of the kidney usually seen in domestic and wild carnivores and observado normalmente em carnívoros domésticos e silvestres e rarely in human beings. This is a report about the parasitism excepcionalmente em seres humanos. Relata-se o parasitismo por D. of D. renale found in the kidney of two roadkill lesser grisons renale em rim de dois furões pequenos (Galictis cuja) encontrados (Galictis cuja) in the North of the state of Santa Catarina, mortos por atropelamento no Norte do estado de Santa Catarina, Brazil. The report of this parasitism in this species is important Brasil. Relatar esse parasitismo nessa espécie é importante, para to complement the records about this native carnivore as a que se possam somar dados relativos a participação deste carnívoro contributor in the epidemiologic chain while host/disseminator nativo na cadeia epidemiológica como hospedeiro/ veiculador desse of this helminth with zoonotic potential. helminto com potencial zoonótico. KEYWORDS: Dioctophyma; wild animal; mustelids; roadkill; PALAVRAS-CHAVE: Dioctophyma; animal silvestre; mustelí- kidney parasitism. deo; atropelamento em rodovia; parasitismo renal. 1Universidade do Contestado (UnC) – Canoinhas (SC), Brazil. *Corresponding author: [email protected] Received on: 04/22/2016. Accepted on: 09/12/2017 Arq. Inst. Biol., v.84, 1-4, e0312016, 2017 1 D. -

Notes on the Distribution, Status, and Research Priorities of Little-Known Small Carnivores in Brazil
Notes on the distribution, status, and research priorities of little-known small carnivores in Brazil Tadeu G. de OLIVEIRA Abstract Ten species of small carnivores occur in Brazil, including four procyonids, four mustelids (excluding otters), and two mephitids. On the IUCN Red List of Threatened Species eight are assessed as Least Concern and two as Data Deficient. The state of knowledge of small carnivores is low compared to other carnivores: they are among the least known of all mammals in Brazil. The current delineation of Bassaricyon and Galictis congeners appears suspect and not based on credible information. Research needs include understanding dis- tributions, ecology and significant evolutionary units, with emphasis on theAmazon Weasel Mustela africana. Keywords: Amazon weasel, Data Deficient, Olingo, Crab-eating Raccoon, Hog-nosed Skunk Notas sobre la distribución, estado y prioridades de investigación de los pequeños carnívoros de Brasil Resumen En Brasil ocurren diez especies de pequeños carnívoros, incluyendo cuatro prociónidos, cuatro mustélidos (excluyendo nutrias) y dos mephitidos. De acuerdo a la Lista Roja de Especies Amenazadas de la UICN, ocho especies son evaluadas como de Baja Preocupación (LC) y dos son consideradas Deficientes de Datos (DD). El estado de conocimiento de los pequeños carnívoros es bajo comparado con otros carnívoros y se encuentran entre los mamíferos menos conocidos de Brasil. La delineación congenérica actual de Bassaricyon y Galictis parece sospechosa y no basada en información confiable. Las necesidades de investigación incluyen el entendimiento de las distribuciones, ecología y unidades evolutivas significativas, con énfasis en la ComadrejaAmazónica Mustela africana. Palabras clave: Comadreja Amazónica, Deficiente de Datos, Mapache Cangrejero, Olingo, Zorrillo Introduction 1999), but recently has been recognised (e.g. -

A New Otter of Giant Size, Siamogale Melilutra Sp. Nov. \(Lutrinae
UCLA UCLA Previously Published Works Title A new otter of giant size, Siamogale melilutra sp. nov. (Lutrinae: Mustelidae: Carnivora), from the latest Miocene Shuitangba site in north-eastern Yunnan, south-western China, and a total- evidence phylogeny of lutrines Permalink https://escholarship.org/uc/item/11b1d0h9 Journal Journal of Systematic Palaeontology, 16(1) ISSN 1477-2019 Authors Wang, X Grohé, C Su, DF et al. Publication Date 2018-01-02 DOI 10.1080/14772019.2016.1267666 Peer reviewed eScholarship.org Powered by the California Digital Library University of California Journal of Systematic Palaeontology ISSN: 1477-2019 (Print) 1478-0941 (Online) Journal homepage: http://www.tandfonline.com/loi/tjsp20 A new otter of giant size, Siamogale melilutra sp. nov. (Lutrinae: Mustelidae: Carnivora), from the latest Miocene Shuitangba site in north- eastern Yunnan, south-western China, and a total- evidence phylogeny of lutrines Xiaoming Wang, Camille Grohé, Denise F. Su, Stuart C. White, Xueping Ji, Jay Kelley, Nina G. Jablonski, Tao Deng, Youshan You & Xin Yang To cite this article: Xiaoming Wang, Camille Grohé, Denise F. Su, Stuart C. White, Xueping Ji, Jay Kelley, Nina G. Jablonski, Tao Deng, Youshan You & Xin Yang (2017): A new otter of giant size, Siamogale melilutra sp. nov. (Lutrinae: Mustelidae: Carnivora), from the latest Miocene Shuitangba site in north-eastern Yunnan, south-western China, and a total-evidence phylogeny of lutrines, Journal of Systematic Palaeontology, DOI: 10.1080/14772019.2016.1267666 To link to this article: http://dx.doi.org/10.1080/14772019.2016.1267666 View supplementary material Published online: 22 Jan 2017. Submit your article to this journal View related articles View Crossmark data Full Terms & Conditions of access and use can be found at http://www.tandfonline.com/action/journalInformation?journalCode=tjsp20 Download by: [UCLA Library] Date: 23 January 2017, At: 00:17 Journal of Systematic Palaeontology, 2017 http://dx.doi.org/10.1080/14772019.2016.1267666 A new otter of giant size, Siamogale melilutra sp. -

Museum of Natural History
p m r- r-' ME FYF-11 - - T r r.- 1. 4,6*. of the FLORIDA MUSEUM OF NATURAL HISTORY THE COMPARATIVE ECOLOGY OF BOBCAT, BLACK BEAR, AND FLORIDA PANTHER IN SOUTH FLORIDA David Steffen Maehr Volume 40, No. 1, pf 1-176 1997 == 46 1ms 34 i " 4 '· 0?1~ I. Al' Ai: *'%, R' I.' I / Em/-.Ail-%- .1/9" . -_____- UNIVERSITY OF FLORIDA GAINESVILLE Numbers of the BULLETIN OF THE FLORIDA MUSEUM OF NATURAL HISTORY am published at irregular intervals Volumes contain about 300 pages and are not necessarily completed in any one calendar year. JOHN F. EISENBERG, EDITOR RICHARD FRANZ CO-EDIWR RHODA J. BRYANT, A£ANAGING EMOR Communications concerning purchase or exchange of the publications and all manuscripts should be addressed to: Managing Editor. Bulletin; Florida Museum of Natural Histoty, University of Florida P. O. Box 117800, Gainesville FL 32611-7800; US.A This journal is printed on recycled paper. ISSN: 0071-6154 CODEN: BF 5BAS Publication date: October 1, 1997 Price: $ 10.00 Frontispiece: Female Florida panther #32 treed by hounds in a laurel oak at the site of her first capture on the Florida Panther National Wildlife Refuge in central Collier County, 3 February 1989. Photograph by David S. Maehr. THE COMPARATIVE ECOLOGY OF BOBCAT, BLACK BEAR, AND FLORIDA PANTHER IN SOUTH FLORIDA David Steffen Maehri ABSTRACT Comparisons of food habits, habitat use, and movements revealed a low probability for competitive interactions among bobcat (Lynx ndia). Florida panther (Puma concotor cooi 1 and black bear (Urns amencanus) in South Florida. All three species preferred upland forests but ©onsumed different foods and utilized the landscape in ways that resulted in ecological separation. -

The 2008 IUCN Red Listings of the World's Small Carnivores
The 2008 IUCN red listings of the world’s small carnivores Jan SCHIPPER¹*, Michael HOFFMANN¹, J. W. DUCKWORTH² and James CONROY³ Abstract The global conservation status of all the world’s mammals was assessed for the 2008 IUCN Red List. Of the 165 species of small carni- vores recognised during the process, two are Extinct (EX), one is Critically Endangered (CR), ten are Endangered (EN), 22 Vulnerable (VU), ten Near Threatened (NT), 15 Data Deficient (DD) and 105 Least Concern. Thus, 22% of the species for which a category was assigned other than DD were assessed as threatened (i.e. CR, EN or VU), as against 25% for mammals as a whole. Among otters, seven (58%) of the 12 species for which a category was assigned were identified as threatened. This reflects their attachment to rivers and other waterbodies, and heavy trade-driven hunting. The IUCN Red List species accounts are living documents to be updated annually, and further information to refine listings is welcome. Keywords: conservation status, Critically Endangered, Data Deficient, Endangered, Extinct, global threat listing, Least Concern, Near Threatened, Vulnerable Introduction dae (skunks and stink-badgers; 12), Mustelidae (weasels, martens, otters, badgers and allies; 59), Nandiniidae (African Palm-civet The IUCN Red List of Threatened Species is the most authorita- Nandinia binotata; one), Prionodontidae ([Asian] linsangs; two), tive resource currently available on the conservation status of the Procyonidae (raccoons, coatis and allies; 14), and Viverridae (civ- world’s biodiversity. In recent years, the overall number of spe- ets, including oyans [= ‘African linsangs’]; 33). The data reported cies included on the IUCN Red List has grown rapidly, largely as on herein are freely and publicly available via the 2008 IUCN Red a result of ongoing global assessment initiatives that have helped List website (www.iucnredlist.org/mammals). -

University of Florida Thesis Or Dissertation Formatting
UNDERSTANDING CARNIVORAN ECOMORPHOLOGY THROUGH DEEP TIME, WITH A CASE STUDY DURING THE CAT-GAP OF FLORIDA By SHARON ELIZABETH HOLTE A DISSERTATION PRESENTED TO THE GRADUATE SCHOOL OF THE UNIVERSITY OF FLORIDA IN PARTIAL FULFILLMENT OF THE REQUIREMENTS FOR THE DEGREE OF DOCTOR OF PHILOSOPHY UNIVERSITY OF FLORIDA 2018 © 2018 Sharon Elizabeth Holte To Dr. Larry, thank you ACKNOWLEDGMENTS I would like to thank my family for encouraging me to pursue my interests. They have always believed in me and never doubted that I would reach my goals. I am eternally grateful to my mentors, Dr. Jim Mead and the late Dr. Larry Agenbroad, who have shaped me as a paleontologist and have provided me to the strength and knowledge to continue to grow as a scientist. I would like to thank my colleagues from the Florida Museum of Natural History who provided insight and open discussion on my research. In particular, I would like to thank Dr. Aldo Rincon for his help in researching procyonids. I am so grateful to Dr. Anne-Claire Fabre; without her understanding of R and knowledge of 3D morphometrics this project would have been an immense struggle. I would also to thank Rachel Short for the late-night work sessions and discussions. I am extremely grateful to my advisor Dr. David Steadman for his comments, feedback, and guidance through my time here at the University of Florida. I also thank my committee, Dr. Bruce MacFadden, Dr. Jon Bloch, Dr. Elizabeth Screaton, for their feedback and encouragement. I am grateful to the geosciences department at East Tennessee State University, the American Museum of Natural History, and the Museum of Comparative Zoology at Harvard for the loans of specimens. -
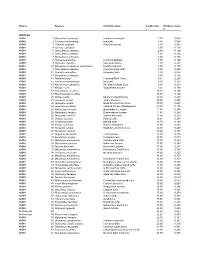
PDF File Containing Table of Lengths and Thicknesses of Turtle Shells And
Source Species Common name length (cm) thickness (cm) L t TURTLES AMNH 1 Sternotherus odoratus common musk turtle 2.30 0.089 AMNH 2 Clemmys muhlenbergi bug turtle 3.80 0.069 AMNH 3 Chersina angulata Angulate tortoise 3.90 0.050 AMNH 4 Testudo carbonera 6.97 0.130 AMNH 5 Sternotherus oderatus 6.99 0.160 AMNH 6 Sternotherus oderatus 7.00 0.165 AMNH 7 Sternotherus oderatus 7.00 0.165 AMNH 8 Homopus areolatus Common padloper 7.95 0.100 AMNH 9 Homopus signatus Speckled tortoise 7.98 0.231 AMNH 10 Kinosternon subrabum steinochneri Florida mud turtle 8.90 0.178 AMNH 11 Sternotherus oderatus Common musk turtle 8.98 0.290 AMNH 12 Chelydra serpentina Snapping turtle 8.98 0.076 AMNH 13 Sternotherus oderatus 9.00 0.168 AMNH 14 Hardella thurgi Crowned River Turtle 9.04 0.263 AMNH 15 Clemmys muhlenbergii Bog turtle 9.09 0.231 AMNH 16 Kinosternon subrubrum The Eastern Mud Turtle 9.10 0.253 AMNH 17 Kinixys crosa hinged-back tortoise 9.34 0.160 AMNH 18 Peamobates oculifers 10.17 0.140 AMNH 19 Peammobates oculifera 10.27 0.140 AMNH 20 Kinixys spekii Speke's hinged tortoise 10.30 0.201 AMNH 21 Terrapene ornata ornate box turtle 10.30 0.406 AMNH 22 Terrapene ornata North American box turtle 10.76 0.257 AMNH 23 Geochelone radiata radiated tortoise (Madagascar) 10.80 0.155 AMNH 24 Malaclemys terrapin diamondback terrapin 11.40 0.295 AMNH 25 Malaclemys terrapin Diamondback terrapin 11.58 0.264 AMNH 26 Terrapene carolina eastern box turtle 11.80 0.259 AMNH 27 Chrysemys picta Painted turtle 12.21 0.267 AMNH 28 Chrysemys picta painted turtle 12.70 0.168 AMNH 29 -

Chewing and Sucking Lice As Parasites of Iviammals and Birds
c.^,y ^r-^ 1 Ag84te DA Chewing and Sucking United States Lice as Parasites of Department of Agriculture IVIammals and Birds Agricultural Research Service Technical Bulletin Number 1849 July 1997 0 jc: United States Department of Agriculture Chewing and Sucking Agricultural Research Service Lice as Parasites of Technical Bulletin Number IVIammals and Birds 1849 July 1997 Manning A. Price and O.H. Graham U3DA, National Agrioultur«! Libmry NAL BIdg 10301 Baltimore Blvd Beltsvjlle, MD 20705-2351 Price (deceased) was professor of entomoiogy, Department of Ento- moiogy, Texas A&iVI University, College Station. Graham (retired) was research leader, USDA-ARS Screwworm Research Laboratory, Tuxtia Gutiérrez, Chiapas, Mexico. ABSTRACT Price, Manning A., and O.H. Graham. 1996. Chewing This publication reports research involving pesticides. It and Sucking Lice as Parasites of Mammals and Birds. does not recommend their use or imply that the uses U.S. Department of Agriculture, Technical Bulletin No. discussed here have been registered. All uses of pesti- 1849, 309 pp. cides must be registered by appropriate state or Federal agencies or both before they can be recommended. In all stages of their development, about 2,500 species of chewing lice are parasites of mammals or birds. While supplies last, single copies of this publication More than 500 species of blood-sucking lice attack may be obtained at no cost from Dr. O.H. Graham, only mammals. This publication emphasizes the most USDA-ARS, P.O. Box 969, Mission, TX 78572. Copies frequently seen genera and species of these lice, of this publication may be purchased from the National including geographic distribution, life history, habitats, Technical Information Service, 5285 Port Royal Road, ecology, host-parasite relationships, and economic Springfield, VA 22161. -

Asian Small-Clawed Otter Husbandry Manual/Health Care -19- Small-Clawed Otters
Introductiion IV--VII Status…………………………………………………………… V SSP Members…………………………………………………... V-VI Management Group Members………………………………….. VI Husbandry Manual Group………………………………………. VI-VII Chapter 1—Nutriitiion & diiet 1--18 Feeding Ecology…………………………………………………. 1 Target Dietary Nutrient Values………………………………….. 2 Food Items Available to Zoos……………………………………. 2 Zoo Diet Summary……………………………………………….. 2-4 Recommendations for Feeding…………………………………… 4-6 Hand Rearing/Infant Diet………………………………………… 6-7 Alternative Diets…………………………………………………. 7-8 Reported Health Problems Associated with Diet………………… 8-9 Future Research Needs…………………………………………… 9 Table 1.1 & 1.2…………………………………………….…….. 10 Table 1.3…………………………………………………………. 11 Table 1.4 ….…………………………………………………….. 12 Table 1.5,1.6 & 1.7……………………………………………… 13 Survey Diet Summary……………………………………..……. 14-15 Appendix 1.1…………………………………………………….. 16 Appendix 1.2…………..…………………………………..…….. 17-18 Chapter 2—Health 19 -- 47 Introduction………………………………………………………. 19 Physiological norms…………………………………………….. 19 Blood baseline values……………………………………………. 19-20 Medical Records………………………………………………….. 21 Identification…………………………………………………….. 21 Preventive Health Care………………………………………….. 21-22 Immunization……………………………………………………. 22 Parasites…………………………………………………………. 23 Pre-shipment examination recommendations…………………… 24 Quarantine………………………………………………………. 24 Control of Reproduction………………………………………… 24-25 Immobilization/anesthesia……………………………………… 25-26 Necropsy Protocol……………………………………………… 26-28 Tissues to be saved…………………………………………….. 29 Table of Contents/Introduction I Diseases