Biodiversity Monitoring Using Environmental DNA: Can It Detect All Fish Species in a Waterbody and Is It Cost Effective for Routine Monitoring?
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

A Global Assessment of Parasite Diversity in Galaxiid Fishes
diversity Article A Global Assessment of Parasite Diversity in Galaxiid Fishes Rachel A. Paterson 1,*, Gustavo P. Viozzi 2, Carlos A. Rauque 2, Verónica R. Flores 2 and Robert Poulin 3 1 The Norwegian Institute for Nature Research, P.O. Box 5685, Torgarden, 7485 Trondheim, Norway 2 Laboratorio de Parasitología, INIBIOMA, CONICET—Universidad Nacional del Comahue, Quintral 1250, San Carlos de Bariloche 8400, Argentina; [email protected] (G.P.V.); [email protected] (C.A.R.); veronicaroxanafl[email protected] (V.R.F.) 3 Department of Zoology, University of Otago, P.O. Box 56, Dunedin 9054, New Zealand; [email protected] * Correspondence: [email protected]; Tel.: +47-481-37-867 Abstract: Free-living species often receive greater conservation attention than the parasites they support, with parasite conservation often being hindered by a lack of parasite biodiversity knowl- edge. This study aimed to determine the current state of knowledge regarding parasites of the Southern Hemisphere freshwater fish family Galaxiidae, in order to identify knowledge gaps to focus future research attention. Specifically, we assessed how galaxiid–parasite knowledge differs among geographic regions in relation to research effort (i.e., number of studies or fish individuals examined, extent of tissue examination, taxonomic resolution), in addition to ecological traits known to influ- ence parasite richness. To date, ~50% of galaxiid species have been examined for parasites, though the majority of studies have focused on single parasite taxa rather than assessing the full diversity of macro- and microparasites. The highest number of parasites were observed from Argentinean galaxiids, and studies in all geographic regions were biased towards the highly abundant and most widely distributed galaxiid species, Galaxias maculatus. -

State of the Fisheries Report 2000-2001 to the Hon
State of the Fisheries Report 2000-2001 To the Hon. Kim Chance MLC Minister for Agriculture, Forestry and Fisheries Sir In accordance with Section 263 of the Fish Resources Management Act 1994, I submit for your information and presentation to Parliament the report State of the Fisheries which forms part of the Annual Report of Fisheries WA for the financial year ending 30 June 2001. Peter P Rogers EXECUTIVE DIRECTOR Edited by Dr J. W. Penn Produced by the Fisheries Research Division based at the WA Marine Research Laboratories Published by the Department of Fisheries 3rd Floor, SGIO Atrium 168 St. Georges Terrace Perth WA 6000 Webiste: http://www.wa.gov.au/westfish Email: [email protected] ISSN 1446 - 5906 (print) ISSN 1446 - 5914 (online) ISSN 1446 - 5922 (CD) Cover photographs: (top) The Department of Fisheries’ new 23m research vessel Naturaliste, constructed and launched in 2001. Photo courtesy Tenix Defence Pty Ltd (shipbuilders). (bottom) Reef habitat at the Houtman Abrolhos Islands. Photo: Clay Bryce State of the Fisheries Report 2000-2001 Glossary of Acronyms yms on ADF Aquaculture Development Fund JASDGDLF Joint Authority Southern Demersal Gillnet and Demersal Longline AFMA Australian Fisheries Management Managed Fishery Authority LML legal minimum length AIMWTMF Abrolhos Islands and Mid West y of Acr Trawl Managed Fishery MAC management advisory committee AQIS Australian Quarantine and MOP mother-of-pearl Inspection Service MSC Marine Stewardship Council Glossar ATSIC Aboriginal and Torres Strait NDSMF Northern Demersal -

Evaluation of Seasonal Dynamics of Fungal DNA Assemblages in a Flow-Regulated Stream in a Restored Forest Using Edna Metabarcoding
bioRxiv preprint doi: https://doi.org/10.1101/2020.12.10.420661; this version posted May 23, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. Evaluation of seasonal dynamics of fungal DNA assemblages in a flow-regulated stream in a restored forest using eDNA metabarcoding Shunsuke Matsuoka1, Yoriko Sugiyama2, Yoshito Shimono3, Masayuki Ushio4,5, Hideyuki Doi1 1. Graduate School of Simulation Studies, University of Hyogo 7-1-28 Minatojima-minamimachi, Chuo-ku, Kobe, 650- 0047, Japan 2. Graduate School of Human and Environmental Studies, Kyoto University, Kyoto 606-8501, Japan 3. Graduate School of Bioresources, Mie University, 1577 Kurima-machiya, Tsu, Mie 514-8507, Japan 4. Hakubi Center, Kyoto University, Yoshida-honmachi, Sakyo-ku, Kyoto 606-8501, Japan 5. Center for Ecological Research, Kyoto University, Hirano 2-509-3, Otsu, Shiga 520-2113, Japan Corresponding author: Shunsuke Matsuoka ([email protected]) Abstract Investigation of seasonal variations in fungal communities is essential for understanding biodiversity and ecosystem functions. However, the conventional sampling method, with substrate removal and high spatial heterogeneity of community compositions, makes surveying the seasonality of fungal communities challenging. Recently, water environmental DNA (eDNA) analysis has been explored for its utility in biodiversity surveys. In this study, we assessed whether the seasonality of fungal communities can be detected by monitoring eDNA in a forest stream. We conducted monthly water sampling in a forest stream over two years and used DNA metabarcoding to identify fungal eDNA. -

Environmental DNA for Wildlife Biology and Biodiversity Monitoring
Review Environmental DNA for wildlife biology and biodiversity monitoring 1,2* 3* 1,4 3 Kristine Bohmann , Alice Evans , M. Thomas P. Gilbert , Gary R. Carvalho , 3 3 5,6 3 Simon Creer , Michael Knapp , Douglas W. Yu , and Mark de Bruyn 1 Centre for GeoGenetics, Natural History Museum of Denmark, University of Copenhagen, Øster Voldgade 5–7, 1350 Copenhagen K, Denmark 2 School of Biological Sciences, University of Bristol, Woodland Road, Bristol BS8 1UG, UK 3 Molecular Ecology and Fisheries Genetics Laboratory, School of Biological Sciences, Deiniol Road, Bangor University, Bangor LL57 2UW, UK 4 Trace and Environmental DNA Laboratory, Department of Environment and Agriculture, Curtin University, Perth, Western Australia 6845, Australia 5 State Key Laboratory of Genetic Resources and Evolution, Kunming Institute of Zoology, Chinese Academy of Sciences, 32 Jiaochang East Road, Kunming, Yunnan 650223, China 6 School of Biological Sciences, University of East Anglia, Norwich Research Park, Norwich, Norfolk NR4 7TJ, UK Extraction and identification of DNA from an environ- technology. Today, science fiction is becoming reality as a mental sample has proven noteworthy recently in growing number of biologists are using eDNA for species detecting and monitoring not only common species, detection and biomonitoring, circumventing, or at least but also those that are endangered, invasive, or elusive. alleviating, the need to sight or sample living organisms. Particular attributes of so-called environmental DNA Such approaches are also accelerating the rate of discovery, (eDNA) analysis render it a potent tool for elucidating because no a priori information about the likely species mechanistic insights in ecological and evolutionary pro- found in a particular environment is required to identify cesses. -

Non-Native Freshwater Fish Management in Biosphere Reserves
ISSN 1989‐8649 Manag. Biolog. Invasions, 2010, 1 Abstract Non‐Native freshwater fish management in Biosphere Reserves The consideration of non‐native freshwater fish species in the Andrea PINO‐DEL‐CARPIO*, Rafael MIRANDA & Jordi PUIG management plans of 18 Biosphere Reserves is evaluated. Additionally, impacts caused by introduced freshwater fish species are described. Introduction, Hypotheses and Reserves was created in 1974 and it Some measures to alleviate the Problems for Management is defined in the Statutory ecological effects of fish species Framework of the World network as introductions are proposed, while Biodiversity, the variety of life on “areas of terrestrial and coastal/ paying attention to local development Earth, is disappearing at an marine ecosystems, or a as well. The introduction of non‐native combination thereof, which are species may have negative conse‐ increasing and unprecedented rate quences for the ecosystems. The (Rockström et al. 2009). This internationally recognized within analysis of the management plans of situation contradicts the 2002 the framework of UNESCO the Reserves confirms that non‐native objective to achieve a significant Programme on Man and the freshwater fish species sometimes are reduction of the current rate of Biosphere (MaB)” (UNESCO 1996). not considered in the action plans of biodiversity loss by 2010 To what extent Biosphere Reserves the area. Biosphere Reserve’s (Convention on Biological Diversity fulfil the aim of biodiversity management plans should consider the 2002). One of the multiple initiatives conservation? In order to make a presence of alien species, with the aim adopted to promote biodiversity first inquiry in this issue, some to preserve biodiversity. -

Hemichromis Bimaculatus (African Jewelfish)
African Jewelfish (Hemichromis bimaculatus) Ecological Risk Screening Summary U.S. Fish and Wildlife Service, April 2011 Revised, September 2018 Web Version, 2/14/2019 Photo: Zhyla. Licensed under CC BY-SA 3.0. Available: https://commons.wikimedia.org/wiki/File:Hemichromis_bimaculatus1.jpg. (September 2018). 1 Native Range and Status in the United States Native Range From Froese and Pauly (2018): “Africa: widely distributed in West Africa, where it is known from most hydrographic basins [Teugels and Thys van den Audenaerde 2003], associated with forested biotopes [Daget and Teugels 1991, Lamboj 2004]. Also reported from coastal basins of Cameroon, Democratic Republic of the Congo and Nile basin [Teugels and Thys van den Audenaerde 1992], but at least its presence in Cameroon is unconfirmed in [Stiassny et al. 2008]. [Lamboj 2004] limits this species to Guinea, Sierra Leone and Liberia.” 1 From Azeroual and Lalèyè (2010): “This species is widely distributed throughout western Africa, but has also been recorded from Algeria to Egypt.” “Northern Africa: Within this region this species is very rare. It used to be caught from the coastal lagoons, especially Lake Mariut (Egypt) and Algeria. Its [sic] found in Tunisia in the wadis of Kebili in the south of Tunisia and in wadis near Chott Melrhir in eastern Algeria (Kraiem, pers. comm.), and Egypt (Wadi El Rayan Lakes).” “Western Africa: It is known from most hydrographic basins in western Africa.” Status in the United States It is not certain if this species is present in the United States, or if records pertain to H. letourneuxi. From NatureServe (2018): “Introduced and established in Dade County, Florida, […] (Nelson 1983).” From Nico et al. -

Croaking Gourami, Trichopsis Vittata (Cuvier, 1831), in Florida, USA
BioInvasions Records (2013) Volume 2, Issue 3: 247–251 Open Access doi: http://dx.doi.org/10.3391/bir.2013.2.3.12 © 2013 The Author(s). Journal compilation © 2013 REABIC Rapid Communication Croaking gourami, Trichopsis vittata (Cuvier, 1831), in Florida, USA Pamela J. Schofield 1* and Darren J. Pecora2 1 US Geological Survey, Southeast Ecological Science Center, 7920 NW 71st Street, Gainesville, FL 32653, USA 2 US Fish and Wildlife Service, Arthur R. Marshall Loxahatchee National Wildlife Refuge, 10216 Lee Road, Boynton Beach, FL 33473, USA E-mail: [email protected] (PJS), [email protected] (DJP) *Corresponding author Received: 8 February 2013 / Accepted: 30 May 2013 / Published online: 1 July 2013 Handling editor: Kit Magellan Abstract The croaking gourami, Trichopsis vittata, is documented from wetland habitats in southern Florida. This species was previously recorded from the same area over 15 years ago, but was considered extirpated. The rediscovery of a reproducing population of this species highlights the dearth of information available regarding the dozens of non-native fishes in Florida, as well as the need for additional research and monitoring. Key words: canal; croaking gourami; cypress swamp; Florida; Loxahatchee; Osphronemidae; Trichopsis vittata was previously considered extirpated (Shafland Introduction et al. 2008a, b), but is now known to be reproducing in a localised area. Dozens of non-native fishes have been introduced into Florida’s inland waterways, via accidental escape, pet releases, or intentional introduction -

Environmental DNA for Improved Detection and Environmental Surveillance of Schistosomiasis
Environmental DNA for improved detection and environmental surveillance of schistosomiasis Mita E. Senguptaa,1, Micaela Hellströmb,c, Henry C. Kariukid, Annette Olsena, Philip F. Thomsenb,e, Helena Mejera, Eske Willerslevb,f,g,h, Mariam T. Mwanjei, Henry Madsena, Thomas K. Kristensena, Anna-Sofie Stensgaardj,2, and Birgitte J. Vennervalda,2 aDepartment of Veterinary and Animal Sciences, University of Copenhagen, DK-1870 Frederiksberg, Denmark; bCentre for GeoGenetics, Natural History Museum of Denmark, University of Copenhagen, DK-1350 Copenhagen K, Denmark; cAquabiota Water Research, SE-115 50, Sweden; dDepartment of Microbiology and Parasitology, Kenya Methodist University, 60200 Meru, Kenya; eDepartment of Bioscience, University of Aarhus, DK-8000 Aarhus C, Denmark; fDepartment of Zoology, University of Cambridge, CB2 3EJ Cambridge, United Kingdom; gHuman Genetics Programme, Wellcome Trust Sanger Institute, Hinxton, SB10 1SA Cambridge, United Kingdom; hDanish Institute for Advanced Study, University of Southern Denmark, DK-5230 Odense M, Denmark; iNeglected Tropical Diseases Unit, Division of Communicable Disease Prevention and Control, Ministry of Health, Nairobi, Kenya; and jCenter for Macroecology, Evolution and Climate, Natural History Museum of Denmark, University of Copenhagen, DK-2100 Copenhagen, Denmark Edited by Andrea Rinaldo, École Polytechnique Fédérale de Lausanne (EPFL), Lausanne, Switzerland, and approved March 18, 2019 (received for review September 21, 2018) Schistosomiasis is a water-based, infectious disease with -

African Jewelfish ( Hemichromis Letourneuxi )
African Jewelfish ( Hemichromis letourneuxi ) Order: Perciformes - Family: Cichlidae - Subfamily: Pseudocrenilabrinae - Tribe: Hemichromini Also known as: Saraba or Nile Jewell Fish Synonyms: Hemichromis rolandi, Hemichromis saharae, Hemichromis bimaculatus saharae, Hemichromis letourneauxi. Type: Freshwater, brackish; benthopelagic; - African Cichlid_Sauvage, 1880 Overview: Described as Hemichromis letourneuxi (Sauvage 1880), but commonly misspelled as H. letourneauxi. Loiselle (1979) revised the genus and provided diagno- ses, photographs, and synonyms for the species. An updated key to the genus was given in Loiselle (1992). Color photographs were provided by Linke and Staeck (1994). Until recently, all published references to introduced populations of this species taken in Florida were incorrectly identified or listed as Hemichromis bimaculatus (Smith-Vaniz, personal communication). Description: Physical Characteristics: Color Form: Max. Size: Approximate size 12-15 см Size 10-12 cm (3.9-4.7") Sexual dimorphism: Diet: Carnivore, Pellet Foods, Flake Foods, Live Foods Lifespan: 5-8 years Reproduction & Spawning: Behavior: . Habitat: Savannah associated species which prospers in a range of lentic habitats that include brackish water lagoons, large lakes and riverine flood plains (Ref. 5644). Occurs near vegetation beds and fringes. Feeds on Caridina and insects. Substrate spawner, ripe and spent fish are common early in the flood season. Origin / Distribution: The African Jewelfish, (Hemichromis letourneuxi) is native to the north and northwestern regions of Africa. Although the species has been present in the canals of south Florida since the 1950s, its geographic range has expanded greatly in recent years and continues to spread throughout south Florida habitats, from Ever- glades National Park to Big Cypress National Park (Loftus and others 2006). -
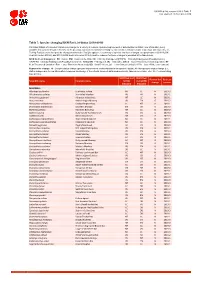
Table 7: Species Changing IUCN Red List Status (2018-2019)
IUCN Red List version 2019-3: Table 7 Last Updated: 10 December 2019 Table 7: Species changing IUCN Red List Status (2018-2019) Published listings of a species' status may change for a variety of reasons (genuine improvement or deterioration in status; new information being available that was not known at the time of the previous assessment; taxonomic changes; corrections to mistakes made in previous assessments, etc. To help Red List users interpret the changes between the Red List updates, a summary of species that have changed category between 2018 (IUCN Red List version 2018-2) and 2019 (IUCN Red List version 2019-3) and the reasons for these changes is provided in the table below. IUCN Red List Categories: EX - Extinct, EW - Extinct in the Wild, CR - Critically Endangered [CR(PE) - Critically Endangered (Possibly Extinct), CR(PEW) - Critically Endangered (Possibly Extinct in the Wild)], EN - Endangered, VU - Vulnerable, LR/cd - Lower Risk/conservation dependent, NT - Near Threatened (includes LR/nt - Lower Risk/near threatened), DD - Data Deficient, LC - Least Concern (includes LR/lc - Lower Risk, least concern). Reasons for change: G - Genuine status change (genuine improvement or deterioration in the species' status); N - Non-genuine status change (i.e., status changes due to new information, improved knowledge of the criteria, incorrect data used previously, taxonomic revision, etc.); E - Previous listing was an Error. IUCN Red List IUCN Red Reason for Red List Scientific name Common name (2018) List (2019) change version Category -

Summary Report of Freshwater Nonindigenous Aquatic Species in U.S
Summary Report of Freshwater Nonindigenous Aquatic Species in U.S. Fish and Wildlife Service Region 4—An Update April 2013 Prepared by: Pam L. Fuller, Amy J. Benson, and Matthew J. Cannister U.S. Geological Survey Southeast Ecological Science Center Gainesville, Florida Prepared for: U.S. Fish and Wildlife Service Southeast Region Atlanta, Georgia Cover Photos: Silver Carp, Hypophthalmichthys molitrix – Auburn University Giant Applesnail, Pomacea maculata – David Knott Straightedge Crayfish, Procambarus hayi – U.S. Forest Service i Table of Contents Table of Contents ...................................................................................................................................... ii List of Figures ............................................................................................................................................ v List of Tables ............................................................................................................................................ vi INTRODUCTION ............................................................................................................................................. 1 Overview of Region 4 Introductions Since 2000 ....................................................................................... 1 Format of Species Accounts ...................................................................................................................... 2 Explanation of Maps ................................................................................................................................ -

DNA Barcoding for Species Identification and Discovery in Diatoms
557_578_MANN.fm Page 557 Mercredi, 8. décembre 2010 8:58 08 Cryptogamie, Algologie, 2010, 31 (4): 557-577 © 2010 Adac. Tous droits réservés DNA barcoding for species identification and discovery in diatoms David G. MANN a* , Shinya SATO a, Rosa TROBAJO b, Pieter VANORMELINGEN c & Caroline SOUFFREAU c a Royal Botanic Garden Edinburgh, Inverleith Row, Edinburgh EH3 5LR, Scotland, UK b Aquatic Ecosystems, Institute for Food and Agricultural Research and Technology (IRTA), Ctra Poble Nou Km 5.5, Sant Carles de la Ràpita, Catalunya, E-43540, Spain c Laboratory of Protistology and Aquatic Ecology, Department of Biology, Ghent University, Krijgslaan 281-S8, B-9000 Gent, Belgium Abstract – Diatoms are the largest group of microalgae, play an enormous role in the biosphere, and have major significance as bioindicators. Traditional identification requires inter alia long training, considerable microscopical skill, and use of a vast and scattered literature. During the life cycle, diatom cells change in size and pattern, often also shape, but the full cycle is known in <1% of described species. Recent evidence shows that there are many pseudocryptic and cryptic species of diatoms, requiring molecular methods for discovery and recognition. These and other factors argue that DNA barcoding would be highly beneficial. It could be ‘strong’, resolving nearly all species, or ‘weak’, resolving mostly species already recognized from light microscopy. Attempts have already been made to identify suitable genes and we evaluate these on the basis of universality and practicality, and ability to discriminate between species in the very few ‘model’ systems offering likely examples of sister-species-pairs. No candidate marker is ideal but LSU rDNA and rbcL may be acceptable, though their discriminatory power is lower than that of some other markers.