Mating-Type Genes of the Anamorphic Fungus Ulocladium Botrytis Affect
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

In Tetrahymena Thermophila Jacek Gaertig,* Manuel A
Acetylation of Lysine 40 in ot-tubulin Is Not Essential in Tetrahymena thermophila Jacek Gaertig,* Manuel A. Cruz, Josephine Bowen, Long Gu, David G. Pennock,* and Martin A. Gorovsky Department of Biology, University of Rochester, Rochester, New York 14627; and *Department of Zoology, Miami University, Oxford, Ohio 45056 Abstract. In Tetrahymena, at least 17 distinct microtu- acetylated tubulin was detectable in these transform- bule structures are assembled from a single primary se- ants using a monoclonal antibody specific for acety- quence type of oL- and 13-tubulin heterodimer, preclud- lated lysine 40. Surprisingly, mutants lacking detectable ing distinctions among microtubular systems based on acetylated tubulin are indistinguishable from wild-type tubulin primary sequence isotypes. Tetrahymena tubu- cells. Thus, acetylation of o~-tubulin at lysine 40 is non- lins also are modified by several types of posttransla- essential in Tetrahymena. In addition, isoelectric focus- tional reactions including acetylation of a-tubulin at ing gel analysis of axonemal tubulin from cells unable lysine 40, a modification found in most eukaryotes. In to acetylate o~-tubulin leads us to conclude that: (a) Tetrahyrnena, axonemal o~-tubulin and numerous other most or all ciliary ot-tubulin is acetylated, (b) other microtubules are acetylated. We completely replaced lysines cannot be acetylated to compensate for loss of the single type of a-tubulin gene in the macronucleus acetylation at lysine 40, and (c) acetylated o~-tubulin with a version encoding arginine instead of lysine 40 molecules in wild-type cells contain one or more addi- and therefore cannot be acetylated at this position. No tional charge-altering modifications. -

Genetics of Polyketide Metabolism in Aspergillus Nidulans
Metabolites 2012, 2, 100-133; doi:10.3390/metabo2010100 OPEN ACCESS metabolites ISSN 2218-1989 www.mdpi.com/journal/metabolites/ Review Genetics of Polyketide Metabolism in Aspergillus nidulans Marie L. Klejnstrup 1, Rasmus J. N. Frandsen 2, Dorte K. Holm 2, Morten T. Nielsen 2, Uffe H. Mortensen 2, Thomas O. Larsen 1 and Jakob B. Nielsen 2,* 1 Department of Systems Biology, Center for Microbial Biotechnology, Technical University of Denmark, Søltofts Plads B221, DK-2800 Kgs. Lyngby, Denmark; E-Mails: [email protected] (M.L.K.); [email protected] (T.O.L) 2 Department of Systems Biology, Center for Microbial Biotechnology, Technical University of Denmark, Søltofts Plads B223, DK-2800 Kgs. Lyngby, Denmark; E-Mails: [email protected] (R.J.N.F.); [email protected] (D.K.H.); [email protected] (M.T.N.); [email protected] (U.H.M.) * Author to whom correspondence should be addressed; E-Mail: [email protected]; Tel.: +45-4525-2657; Fax: +45-4588-4148. Received: 1 November 2011; in revised form: 23 December 2011 / Accepted: 17 January 2012 / Published: 30 January 2012 Abstract: Secondary metabolites are small molecules that show large structural diversity and a broad range of bioactivities. Some metabolites are attractive as drugs or pigments while others act as harmful mycotoxins. Filamentous fungi have the capacity to produce a wide array of secondary metabolites including polyketides. The majority of genes required for production of these metabolites are mostly organized in gene clusters, which often are silent or barely expressed under laboratory conditions, making discovery and analysis difficult. -

An Unusual Haemoid Fungi: Ulocladium, As a Cause Of
Volume 2 Number 2 (June 2010) 95-97 CASE Report An unusual phaeoid fungi: Ulocladium, as a cause of chronic allergic fungal sinusitis Kaur R, Wadhwa A1,Gulati A2, Agrawal AK2 1Department of Microbiology. 2Department of Otorhinolaryngology, Maulana Azad Medical College, New Delhi, India. Received: April 2010, Accepted: May 2010. ABSTRACT Allergic fungal sinusitis (AFS) has been recognized as an important cause of chronic sinusitis commonly caused by Aspergillus spp. and various dematiaceous fungi like Bipolaris, Alternaria, Curvalaria, and etc. Ulocladium botrytis is a non pathogenic environmental dematiaceous fungi, which has been recently described as a human pathogen. Ulocladium has never been associated with allergic fungal sinusitis but it was identified as an etiological agent of AFS in a 35 year old immunocompetent female patient presenting with chronic nasal obstruction of several months duration to our hospital. The patient underwent FESS and the excised polyps revealed Ulocladium as the causative fungal agent. Keywords: Ulocladium, Phaeoid Fungi, Chronic Sinusitis. INTRODUCTION CASE REPORT Chronic allergic sinusitis is a common condition A 35 year old female patient presented to the ENT responsible for the development of nasal polyps, out patient department of the Lok Nayak Hospital, described as abnormal lesions that emanate from Delhi, with chronic nasal obstruction, excessive any portion of the nasal mucosa or Para nasal sneezing, nasal discharge and frontal headache since sinuses. They are commonly located in the middle several months. Nasal obstruction was of gradual meatus and ethmoid sinus and are present in onset, non progressive, more on the left side than 1-4% of the population (1). -

Molecular Systematics of the Marine Dothideomycetes
available online at www.studiesinmycology.org StudieS in Mycology 64: 155–173. 2009. doi:10.3114/sim.2009.64.09 Molecular systematics of the marine Dothideomycetes S. Suetrong1, 2, C.L. Schoch3, J.W. Spatafora4, J. Kohlmeyer5, B. Volkmann-Kohlmeyer5, J. Sakayaroj2, S. Phongpaichit1, K. Tanaka6, K. Hirayama6 and E.B.G. Jones2* 1Department of Microbiology, Faculty of Science, Prince of Songkla University, Hat Yai, Songkhla, 90112, Thailand; 2Bioresources Technology Unit, National Center for Genetic Engineering and Biotechnology (BIOTEC), 113 Thailand Science Park, Paholyothin Road, Khlong 1, Khlong Luang, Pathum Thani, 12120, Thailand; 3National Center for Biothechnology Information, National Library of Medicine, National Institutes of Health, 45 Center Drive, MSC 6510, Bethesda, Maryland 20892-6510, U.S.A.; 4Department of Botany and Plant Pathology, Oregon State University, Corvallis, Oregon, 97331, U.S.A.; 5Institute of Marine Sciences, University of North Carolina at Chapel Hill, Morehead City, North Carolina 28557, U.S.A.; 6Faculty of Agriculture & Life Sciences, Hirosaki University, Bunkyo-cho 3, Hirosaki, Aomori 036-8561, Japan *Correspondence: E.B. Gareth Jones, [email protected] Abstract: Phylogenetic analyses of four nuclear genes, namely the large and small subunits of the nuclear ribosomal RNA, transcription elongation factor 1-alpha and the second largest RNA polymerase II subunit, established that the ecological group of marine bitunicate ascomycetes has representatives in the orders Capnodiales, Hysteriales, Jahnulales, Mytilinidiales, Patellariales and Pleosporales. Most of the fungi sequenced were intertidal mangrove taxa and belong to members of 12 families in the Pleosporales: Aigialaceae, Didymellaceae, Leptosphaeriaceae, Lenthitheciaceae, Lophiostomataceae, Massarinaceae, Montagnulaceae, Morosphaeriaceae, Phaeosphaeriaceae, Pleosporaceae, Testudinaceae and Trematosphaeriaceae. Two new families are described: Aigialaceae and Morosphaeriaceae, and three new genera proposed: Halomassarina, Morosphaeria and Rimora. -

Thesis FINAL PRINT
Preface This study was conducted at the Department of Chemistry, Biotechnology and Food Science (IKBM), Norwegian University of Life Sciences (UMB) during November 2009 to November 2010. My supervisors were Professor Dr Arne Tronsmo and PhD student Md. Hafizur Rahman, Department of IKBM, UMB. I express my sincere appreciation and gratitude to Professor Arne Tronsmo and Dr. Linda Gordon Hjeljord for their dynamic guidance throughout the period of the study, constant encouragement, constructive criticism and valuable suggestion during preparation of the thesis. I also want to thank Md. Hafizur Rahman for his guidance during my research work. Moreover I express my sincere gratitude to Grethe Kobro and Else Maria Aasen and all other staffs and workers at IKBM, UMB for their helpful co-operation to complete the research work in the laboratory. Last but not the least; I feel the heartiest indebtedness to Sabine and my family members for their patient inspirations, sacrifices and never ending encouragement. Ås, January 2011 Latifur Rahman Shovan i Abstract This thesis has been focused on methods to control diseases caused by Botrytis cinerea. B. cinerea causes grey mould disease of strawberry and chickpea, as well as many other plants. The fungal isolates used were isolated from chickpea leaf (Gazipur, Bangladesh) or obtained from the Norwegian culture collections of Bioforsk (Ås) and IKBM (UMB). Both morphological and molecular characterization helped to identify the fungal isolates as Botrytis cinerea (B. cinerea 101 and B. cinerea-BD), Trichoderma atroviride, T. asperellum Alternaria brassicicola, and Mucor piriformis. The identity of one fungal isolate, which was obtained from the culture collection of Bioforsk under the name Microdochium majus, could not be confirmed in this study. -
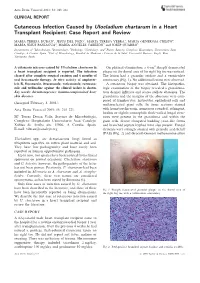
Cutaneous Infection Caused by Ulocladium Chartarum in a Heart Transplant Recipient: Case Report and Review
Acta Derm Venereol 2003; 83: 218–221 CLINICAL REPORT Cutaneous Infection Caused by Ulocladium chartarum in a Heart Transplant Recipient: Case Report and Review MARI´A TERESA DURA´ N1, JESU´ S DEL POZO2, MARI´A TERESA YEBRA3, MARI´A GENEROSA CRESPO4, MARI´A JESU´ S PANIAGUA4, MARI´A ANGELES CABEZO´ N5 and JOSEP GUARRO6 Departments of 1Microbiology, 2Dermatology, 3Pathology, 4Cardiology, and 5Plastic Surgery, Complexo Hospitalario Universitario Juan Canalejo, A Corun˜a, Spain, 6Unit of Microbiology, Facultat de Medicina i Cie`ncies de la Salut, Universitat Rovira i Virgili, Reus, Tarragona, Spain A cutaneous mycoses caused by Ulocladium chartarum in On physical examination, a 6-cm2 sharply demarcated a heart transplant recipient is reported. The infection plaque on the dorsal area of his right big toe was noticed. cleared after complete surgical excision and 6 months of The lesion had a granular surface and a vermiculate oral itraconazole therapy. In vitro activity of amphoter- consistency (Fig. 1). No additional lesions were observed. icin B, fluconazole, itraconazole, voriconazole, ravucona- A cutaneous biopsy was obtained. The histopatho- zole and terbinafine against the clinical isolate is shown. logic examination of the biopsy revealed a granuloma- Key words: dermatomycoses; immunocompromised host; tous dermal infiltrate and scarce stellate abscesses. The skin diseases. granuloma and the margins of the abscesses were com- posed of lymphocytes, histiocytes, epithelioid cells and (Accepted February 3, 2003.) multinucleated giant cells. In tissue sections stained Acta Derm Venereol 2003; 83: 218–221. with hematoxylin-eosin, numerous rounded, refringent, hyaline or slightly eosinophilic thick-walled fungal struc- Ma Teresa Dura´n Valle, Servicio de Microbiologı´a, tures were present in the granuloma and within the Complexo Hospitalario Universitario Juan Canalejo, giant cells. -

Neutrophil Phagocyte Oxidase Activity Controls Invasive Fungal Growth and Inflammation in Zebrafish Taylor J
© 2019. Published by The Company of Biologists Ltd | Journal of Cell Science (2020) 133, jcs236539. doi:10.1242/jcs.236539 RESEARCH ARTICLE Special Issue: Cell Biology of the Immune System Neutrophil phagocyte oxidase activity controls invasive fungal growth and inflammation in zebrafish Taylor J. Schoen1,2, Emily E. Rosowski1,*, Benjamin P. Knox1, David Bennin1, Nancy P. Keller1,3 and Anna Huttenlocher1,4,‡ ABSTRACT aspergillosis is increased in people with inherited Neutrophils are primary phagocytes of the innate immune system that immunodeficiency or medically-induced immunosuppression that generate reactive oxygen species (ROS) and mediate host defense. impairs innate immune cell function, especially those with Deficient phagocyte NADPH oxidase (PHOX) function leads to neutropenia, i.e. neutrophil deficiency (King et al., 2016; Segal chronic granulomatous disease (CGD) that is characterized by et al., 2010). Relative to other inherited immune disorders, CGD is invasive infections, including those by the generally non-pathogenic the most significant predisposing condition for developing invasive fungus Aspergillus nidulans. The role of neutrophil ROS in this aspergillosis (Blumental et al., 2011), and invasive aspergillosis is specific host–pathogen interaction remains unclear. Here, we exploit responsible for many of the infection-related mortalities of CGD the optical transparency of zebrafish to image the effects of neutrophil patients (Henriet et al., 2012; Marciano et al., 2015), with ROS on invasive fungal growth and neutrophil behavior in response to A. fumigatus as the primary causative agent (Marciano et al., Aspergillus nidulans. In a wild-type host, A. nidulans germinates 2015). Despite its rare occurrence in other immunocompromised rapidly and elicits a robust inflammatory response with efficient fungal populations, A. -

Novel Dematiaceous Hyphomycetes
STUDIES IN MYCOLOGY 50: 109–118. 2004. Novel dematiaceous hyphomycetes * Emory G. Simmons 717 Thornwood Road, Crawfordsville, Indiana 47933, U.S.A. *Correspondence: Emory G. Simmons, [email protected] Abstract: Novel taxa are described as Alternaria simsimi isolated from Sesamum indicum, distinguishing it from Alternaria sesami; Embellisia lolii from Lolium perenne; Nimbya perpunctulata from Alternanthera philoxeroides; Ulocladium arbores- cens from Pistacia vera; and Stemphylium symphyti from Symphytum × uplandicum. Taxonomic novelties: Alternaria simsimi E.G. Simmons sp. nov., Embellisia lolii E.G. Simmons & C.F. Hill sp. nov., Nimbya perpunctulata E.G. Simmons sp. nov., Stemphylium symphyti E.G. Simmons sp. nov., Ulocladium arborescens E.G. Simmons sp. nov. Key words: Alternaria, Embellisia, Nimbya, Ulocladium, Stemphylium, anamorphs, Sesamum, Lolium, Alternanthera, Pistacia, Symphytum. INTRODUCTION described and discussed on the basis of examination of colonies at 50× magnification and, at higher magnifi- It is a pleasure to be able to offer several new taxa in cations, of sporulation elements sampled at 5–7 d after honour of the centenary observances of the Centraal- inoculation. Each isolate or specimen discussed is bureau voor Schimmelcultures. These are dedicated to identified by a unique record/research number in the mycologist colleagues who were among my first format E.G.S. 00.000. associates during my earliest visits to CBS in Baarn, beginning in the late 1950s: Agathe van Beverwijk, Gerharda Bunschoten, G.A. de Vries, Amelia Stolk, TAXONOMIC PART Beatrice Schol-Schwarz, Maria Schipper, and Annie van der Plaats-Niterink. A novel taxon is presented for Alternaria: three species on sesame, two as patho- each of the five hyphomycetous genera that have been gens the major focus of my taxonomic work over the past Alternaria isolates from invaded tissues of Sesamum 50+ years. -

Monograph on Dematiaceous Fungi
Monograph On Dematiaceous fungi A guide for description of dematiaceous fungi fungi of medical importance, diseases caused by them, diagnosis and treatment By Mohamed Refai and Heidy Abo El-Yazid Department of Microbiology, Faculty of Veterinary Medicine, Cairo University 2014 1 Preface The first time I saw cultures of dematiaceous fungi was in the laboratory of Prof. Seeliger in Bonn, 1962, when I attended a practical course on moulds for one week. Then I handled myself several cultures of black fungi, as contaminants in Mycology Laboratory of Prof. Rieth, 1963-1964, in Hamburg. When I visited Prof. DE Varies in Baarn, 1963. I was fascinated by the tremendous number of moulds in the Centraalbureau voor Schimmelcultures, Baarn, Netherlands. On the other hand, I was proud, that El-Sheikh Mahgoub, a Colleague from Sundan, wrote an internationally well-known book on mycetoma. I have never seen cases of dematiaceous fungal infections in Egypt, therefore, I was very happy, when I saw the collection of mycetoma cases reported in Egypt by the eminent Egyptian Mycologist, Prof. Dr Mohamed Taha, Zagazig University. To all these prominent mycologists I dedicate this monograph. Prof. Dr. Mohamed Refai, 1.5.2014 Heinz Seeliger Heinz Rieth Gerard de Vries, El-Sheikh Mahgoub Mohamed Taha 2 Contents 1. Introduction 4 2. 30. The genus Rhinocladiella 83 2. Description of dematiaceous 6 2. 31. The genus Scedosporium 86 fungi 2. 1. The genus Alternaria 6 2. 32. The genus Scytalidium 89 2.2. The genus Aurobasidium 11 2.33. The genus Stachybotrys 91 2.3. The genus Bipolaris 16 2. -

PERSOONIAL R Eflections
Persoonia 23, 2009: 177–208 www.persoonia.org doi:10.3767/003158509X482951 PERSOONIAL R eflections Editorial: Celebrating 50 years of Fungal Biodiversity Research The year 2009 represents the 50th anniversary of Persoonia as the message that without fungi as basal link in the food chain, an international journal of mycology. Since 2008, Persoonia is there will be no biodiversity at all. a full-colour, Open Access journal, and from 2009 onwards, will May the Fungi be with you! also appear in PubMed, which we believe will give our authors even more exposure than that presently achieved via the two Editors-in-Chief: independent online websites, www.IngentaConnect.com, and Prof. dr PW Crous www.persoonia.org. The enclosed free poster depicts the 50 CBS Fungal Biodiversity Centre, Uppsalalaan 8, 3584 CT most beautiful fungi published throughout the year. We hope Utrecht, The Netherlands. that the poster acts as further encouragement for students and mycologists to describe and help protect our planet’s fungal Dr ME Noordeloos biodiversity. As 2010 is the international year of biodiversity, we National Herbarium of the Netherlands, Leiden University urge you to prominently display this poster, and help distribute branch, P.O. Box 9514, 2300 RA Leiden, The Netherlands. Book Reviews Mu«enko W, Majewski T, Ruszkiewicz- The Cryphonectriaceae include some Michalska M (eds). 2008. A preliminary of the most important tree pathogens checklist of micromycetes in Poland. in the world. Over the years I have Biodiversity of Poland, Vol. 9. Pp. personally helped collect populations 752; soft cover. Price 74 €. W. Szafer of some species in Africa and South Institute of Botany, Polish Academy America, and have witnessed the of Sciences, Lubicz, Kraków, Poland. -

FGN 46 Aspergillus Bibliography
Fungal Genetics Reports Volume 46 Article 16 FGN 46 Aspergillus Bibliography John Clutterbuck Follow this and additional works at: https://newprairiepress.org/fgr This work is licensed under a Creative Commons Attribution-Share Alike 4.0 License. Recommended Citation Clutterbuck, J. (1999) "FGN 46 Aspergillus Bibliography," Fungal Genetics Reports: Vol. 46, Article 16. https://doi.org/10.4148/1941-4765.1244 This Bibliography is brought to you for free and open access by New Prairie Press. It has been accepted for inclusion in Fungal Genetics Reports by an authorized administrator of New Prairie Press. For more information, please contact [email protected]. FGN 46 Aspergillus Bibliography Abstract This bibliography attempts to cover genetical and biochemical publications on Aspergillus nidulans and also includes selected references to related species and topics. This bibliography is available in Fungal Genetics Reports: https://newprairiepress.org/fgr/vol46/iss1/16 Clutterbuck: FGN 46 Aspergillus Bibliography FGN 46 Aspergillus Bibliography This bibliography attempts to cover genetical and biochemical publications on Aspergillus nidulans and also includes selected references to related species and topics. I would be grateful for publication lists and reprints , especially for papers in books and less readily available periodicals. Entries have been checked as far as possible, but please tell me of any errors. Authors are requested to send a copy of each publication to the FGSC. John Clutterbuck View the Aspergillus Keyword index View the Aspergillus Author index 1. Aleksenko, A. & Ivanova, L. 1998 In vivo linearization and autonomous replication of plasmids containing human telomeric DNA in Aspergillus nidulans. Mol. Gen. Genet. 260: 159- 164 2. -

Functional Characterization of Aspergillus Nidulans Homologues of Saccharomyces Cerevisiae Spa2 and Bud6
University of Nebraska - Lincoln DigitalCommons@University of Nebraska - Lincoln Papers in Plant Pathology Plant Pathology Department June 2006 Functional Characterization of Aspergillus nidulans Homologues of Saccharomyces cerevisiae Spa2 and Bud6 Aleksandra Virag University of Nebraska-Lincoln, [email protected] Steven D. Harris University of Nebraska-Lincoln, [email protected] Follow this and additional works at: https://digitalcommons.unl.edu/plantpathpapers Part of the Plant Pathology Commons Virag, Aleksandra and Harris, Steven D., "Functional Characterization of Aspergillus nidulans Homologues of Saccharomyces cerevisiae Spa2 and Bud6" (2006). Papers in Plant Pathology. 48. https://digitalcommons.unl.edu/plantpathpapers/48 This Article is brought to you for free and open access by the Plant Pathology Department at DigitalCommons@University of Nebraska - Lincoln. It has been accepted for inclusion in Papers in Plant Pathology by an authorized administrator of DigitalCommons@University of Nebraska - Lincoln. EUKARYOTIC CELL, June 2006, p. 881–895 Vol. 5, No. 6 1535-9778/06/$08.00ϩ0 doi:10.1128/EC.00036-06 Copyright © 2006, American Society for Microbiology. All Rights Reserved. Functional Characterization of Aspergillus nidulans Homologues of Saccharomyces cerevisiae Spa2 and Bud6 Aleksandra Virag and Steven D. Harris* Plant Science Initiative and Department of Plant Pathology, University of Nebraska, Lincoln, Nebraska Received 6 February 2006/Accepted 12 April 2006 The importance of polarized growth for fungi has elicited significant effort directed at better understanding underlying mechanisms of polarization, with a focus on yeast systems. At sites of tip growth, multiple protein complexes assemble and coordinate to ensure that incoming building material reaches the appropriate destination sites, and polarized growth is maintained.