OR2G2 (Human) Recombinant Protein (P01)
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

WO 2019/068007 Al Figure 2
(12) INTERNATIONAL APPLICATION PUBLISHED UNDER THE PATENT COOPERATION TREATY (PCT) (19) World Intellectual Property Organization I International Bureau (10) International Publication Number (43) International Publication Date WO 2019/068007 Al 04 April 2019 (04.04.2019) W 1P O PCT (51) International Patent Classification: (72) Inventors; and C12N 15/10 (2006.01) C07K 16/28 (2006.01) (71) Applicants: GROSS, Gideon [EVIL]; IE-1-5 Address C12N 5/10 (2006.0 1) C12Q 1/6809 (20 18.0 1) M.P. Korazim, 1292200 Moshav Almagor (IL). GIBSON, C07K 14/705 (2006.01) A61P 35/00 (2006.01) Will [US/US]; c/o ImmPACT-Bio Ltd., 2 Ilian Ramon St., C07K 14/725 (2006.01) P.O. Box 4044, 7403635 Ness Ziona (TL). DAHARY, Dvir [EilL]; c/o ImmPACT-Bio Ltd., 2 Ilian Ramon St., P.O. (21) International Application Number: Box 4044, 7403635 Ness Ziona (IL). BEIMAN, Merav PCT/US2018/053583 [EilL]; c/o ImmPACT-Bio Ltd., 2 Ilian Ramon St., P.O. (22) International Filing Date: Box 4044, 7403635 Ness Ziona (E.). 28 September 2018 (28.09.2018) (74) Agent: MACDOUGALL, Christina, A. et al; Morgan, (25) Filing Language: English Lewis & Bockius LLP, One Market, Spear Tower, SanFran- cisco, CA 94105 (US). (26) Publication Language: English (81) Designated States (unless otherwise indicated, for every (30) Priority Data: kind of national protection available): AE, AG, AL, AM, 62/564,454 28 September 2017 (28.09.2017) US AO, AT, AU, AZ, BA, BB, BG, BH, BN, BR, BW, BY, BZ, 62/649,429 28 March 2018 (28.03.2018) US CA, CH, CL, CN, CO, CR, CU, CZ, DE, DJ, DK, DM, DO, (71) Applicant: IMMP ACT-BIO LTD. -

Predicting Human Olfactory Perception from Activities of Odorant Receptors
iScience ll OPEN ACCESS Article Predicting Human Olfactory Perception from Activities of Odorant Receptors Joel Kowalewski, Anandasankar Ray [email protected] odor perception HIGHLIGHTS Machine learning predicted activity of 34 human ORs for ~0.5 million chemicals chemical structure Activities of human ORs predicts OR activity could predict odor character using machine learning Few OR activities were needed to optimize r predictions of each odor e t c percept a AI r a odorant activates mul- h Behavior predictions in c Drosophila also need few r tiple ORs o olfactory receptor d o activities ts ic ed pr ity tiv ac OR Kowalewski & Ray, iScience 23, 101361 August 21, 2020 ª 2020 The Author(s). https://doi.org/10.1016/ j.isci.2020.101361 iScience ll OPEN ACCESS Article Predicting Human Olfactory Perception from Activities of Odorant Receptors Joel Kowalewski1 and Anandasankar Ray1,2,3,* SUMMARY Odor perception in humans is initiated by activation of odorant receptors (ORs) in the nose. However, the ORs linked to specific olfactory percepts are unknown, unlike in vision or taste where receptors are linked to perception of different colors and tastes. The large family of ORs (~400) and multiple receptors activated by an odorant present serious challenges. Here, we first use machine learning to screen ~0.5 million compounds for new ligands and identify enriched structural motifs for ligands of 34 human ORs. We next demonstrate that the activity of ORs successfully predicts many of the 146 different perceptual qualities of chem- icals. Although chemical features have been used to model odor percepts, we show that biologically relevant OR activity is often superior. -

Sean Raspet – Molecules
1. Commercial name: Fructaplex© IUPAC Name: 2-(3,3-dimethylcyclohexyl)-2,5,5-trimethyl-1,3-dioxane SMILES: CC1(C)CCCC(C1)C2(C)OCC(C)(C)CO2 Molecular weight: 240.39 g/mol Volume (cubic Angstroems): 258.88 Atoms number (non-hydrogen): 17 miLogP: 4.43 Structure: Biological Properties: Predicted Druglikenessi: GPCR ligand -0.23 Ion channel modulator -0.03 Kinase inhibitor -0.6 Nuclear receptor ligand 0.15 Protease inhibitor -0.28 Enzyme inhibitor 0.15 Commercial name: Fructaplex© IUPAC Name: 2-(3,3-dimethylcyclohexyl)-2,5,5-trimethyl-1,3-dioxane SMILES: CC1(C)CCCC(C1)C2(C)OCC(C)(C)CO2 Predicted Olfactory Receptor Activityii: OR2L13 83.715% OR1G1 82.761% OR10J5 80.569% OR2W1 78.180% OR7A2 77.696% 2. Commercial name: Sylvoxime© IUPAC Name: N-[4-(1-ethoxyethenyl)-3,3,5,5tetramethylcyclohexylidene]hydroxylamine SMILES: CCOC(=C)C1C(C)(C)CC(CC1(C)C)=NO Molecular weight: 239.36 Volume (cubic Angstroems): 252.83 Atoms number (non-hydrogen): 17 miLogP: 4.33 Structure: Biological Properties: Predicted Druglikeness: GPCR ligand -0.6 Ion channel modulator -0.41 Kinase inhibitor -0.93 Nuclear receptor ligand -0.17 Protease inhibitor -0.39 Enzyme inhibitor 0.01 Commercial name: Sylvoxime© IUPAC Name: N-[4-(1-ethoxyethenyl)-3,3,5,5tetramethylcyclohexylidene]hydroxylamine SMILES: CCOC(=C)C1C(C)(C)CC(CC1(C)C)=NO Predicted Olfactory Receptor Activity: OR52D1 71.900% OR1G1 70.394% 0R52I2 70.392% OR52I1 70.390% OR2Y1 70.378% 3. Commercial name: Hyperflor© IUPAC Name: 2-benzyl-1,3-dioxan-5-one SMILES: O=C1COC(CC2=CC=CC=C2)OC1 Molecular weight: 192.21 g/mol Volume -
Quantitative-PCR Validation of 154 Genomic Segments Called As Cnvs in Five Replicat
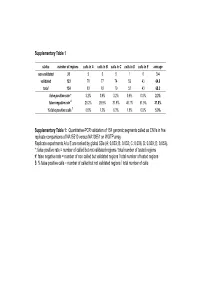
Supplementary Table 1 status number of regions calls in A calls in B calls in C calls in D calls in E average non validated 31 5 6 5 1 0 3.4 validated 123 78 77 74 52 43 64.8 total 154 83 83 79 53 43 68.2 false positive rate * 3.2% 3.9% 3.2% 0.6% 0.0% 2.2% false negative rate # 29.2% 29.9% 31.8% 46.1% 51.9% 37.8% % false positive calls $ 6.0% 7.2% 6.3% 1.9% 0.0% 5.0% Supplementary Table 1: Quantitative-PCR validation of 154 genomic segments called as CNVs in five replicate comparisons of NA15510 versus NA10851 on WGTP array Replicate experiments A to E are ranked by global SDe (A: 0.033; B: 0.033; C: 0.036; D: 0.039; E: 0.053). *: false positive rate = number of called but not validated regions / total number of tested regions #: false negative rate = number of non called but validated regions / total number of tested regions $: % false positive calls = number of called but not validated regions / total number of calls False positive estimates for 500K EA CNV calls Total Rep1 Rep2 Rep3 Avg (unique) Validated 33 28 32 31 38 Not validated 2 2 2 2 5 Total 35 30 34 33 43 % False positive 5.71% 6.67% 5.88% 6.09% - % False negative 13.16% 26.32% 15.79% 18.42% - Supplementary Table 2A : Quantitative PCR validation of 43 unique CNV regions called as CNVs in three replicate comparisons of NA15510 versus NA10851 using the 500K EA array. -

Supplemental Table 1 (S1). the Chromosomal Regions and Genes Exhibiting Loss of Heterozygosity That Were Shared Between the HLRCC-Rccs of Both Patients 1 and 2
BMJ Publishing Group Limited (BMJ) disclaims all liability and responsibility arising from any reliance Supplemental material placed on this supplemental material which has been supplied by the author(s) J Clin Pathol Supplemental Table 1 (S1). The chromosomal regions and genes exhibiting loss of heterozygosity that were shared between the HLRCC-RCCs of both Patients 1 and 2. Chromosome Position Genes 1 p13.1 ATP1A1, ATP1A1-AS1, LOC101929023, CD58, IGSF3, MIR320B1, C1orf137, CD2, PTGFRN, CD101, LOC101929099 1 p21.1* COL11A1, LOC101928436, RNPC3, AMY2B, ACTG1P4, AMY2A, AMY1A, AMY1C, AMY1B 1 p36.11 LOC101928728, ARID1A, PIGV, ZDHHC18, SFN, GPN2, GPATCH3, NR0B2, NUDC, KDF1, TRNP1, FAM46B, SLC9A1, WDTC1, TMEM222, ACTG1P20, SYTL1, MAP3K6, FCN3, CD164L2, GPR3, WASF2, AHDC1, FGR, IFI6 1 q21.3* KCNN3, PMVK, PBXIP1, PYGO2, LOC101928120, SHC1, CKS1B, MIR4258, FLAD1, LENEP, ZBTB7B, DCST2, DCST1, LOC100505666, ADAM15, EFNA4, EFNA3, EFNA1, SLC50A1, DPM3, KRTCAP2, TRIM46, MUC1, MIR92B, THBS3, MTX1, GBAP1, GBA, FAM189B, SCAMP3, CLK2, HCN3, PKLR, FDPS, RUSC1-AS1, RUSC1, ASH1L, MIR555, POU5F1P4, ASH1L-AS1, MSTO1, MSTO2P, YY1AP1, SCARNA26A, DAP3, GON4L, SCARNA26B, SYT11, RIT1, KIAA0907, SNORA80E, SCARNA4, RXFP4, ARHGEF2, MIR6738, SSR2, UBQLN4, LAMTOR2, RAB25, MEX3A, LMNA, SEMA4A, SLC25A44, PMF1, PMF1-BGLAP 1 q24.2–44* LOC101928650, GORAB, PRRX1, MROH9, FMO3, MIR1295A, MIR1295B, FMO6P, FMO2, FMO1, FMO4, TOP1P1, PRRC2C, MYOC, VAMP4, METTL13, DNM3, DNM3-IT1, DNM3OS, MIR214, MIR3120, MIR199A2, C1orf105, PIGC, SUCO, FASLG, TNFSF18, TNFSF4, LOC100506023, LOC101928673, -

Supplementary Table 1
Supplementary Table 1. List of genes that encode proteins contianing cell surface epitopes and are represented on Agilent human 1A V2 microarray chip (2,177 genes) Agilent Probe ID Gene Symbol GenBank ID UniGene ID A_23_P103803 FCRH3 AF459027 Hs.292449 A_23_P104811 TREH AB000824 Hs.129712 A_23_P105100 IFITM2 X57351 Hs.174195 A_23_P107036 C17orf35 X51804 Hs.514009 A_23_P110736 C9 BC020721 Hs.1290 A_23_P111826 SPAM1 NM_003117 Hs.121494 A_23_P119533 EFNA2 AJ007292 No-Data A_23_P120105 KCNS3 BC004987 Hs.414489 A_23_P128195 HEM1 NM_005337 Hs.182014 A_23_P129332 PKD1L2 BC014157 Hs.413525 A_23_P130203 SYNGR2 AJ002308 Hs.464210 A_23_P132700 TDGF1 X14253 Hs.385870 A_23_P1331 COL13A1 NM_005203 Hs.211933 A_23_P138125 TOSO BC006401 Hs.58831 A_23_P142830 PLA2R1 U17033 Hs.410477 A_23_P146506 GOLPH2 AF236056 Hs.494337 A_23_P149569 MG29 No-Data No-Data A_23_P150590 SLC22A9 NM_080866 Hs.502772 A_23_P151166 MGC15619 BC009731 Hs.334637 A_23_P152620 TNFSF13 NM_172089 Hs.54673 A_23_P153986 KCNJ3 U39196 No-Data A_23_P154855 KCNE1 NM_000219 Hs.121495 A_23_P157380 KCTD7 AK056631 Hs.520914 A_23_P157428 SLC10A5 AK095808 Hs.531449 A_23_P160159 SLC2A5 BC001820 Hs.530003 A_23_P162162 KCTD14 NM_023930 Hs.17296 A_23_P162668 CPM BC022276 Hs.334873 A_23_P164341 VAMP2 AL050223 Hs.25348 A_23_P165394 SLC30A6 NM_017964 Hs.552598 A_23_P167276 PAQR3 AK055774 Hs.368305 A_23_P170636 KCNH8 AY053503 Hs.475656 A_23_P170736 MMP17 NM_016155 Hs.159581 A_23_P170959 LMLN NM_033029 Hs.518540 A_23_P19042 GABRB2 NM_021911 Hs.87083 A_23_P200361 CLCN6 X83378 Hs.193043 A_23_P200710 PIK3C2B -

Amino Acid Sequences Directed Against Cxcr4 And
(19) TZZ ¥¥_T (11) EP 2 285 833 B1 (12) EUROPEAN PATENT SPECIFICATION (45) Date of publication and mention (51) Int Cl.: of the grant of the patent: C07K 16/28 (2006.01) A61K 39/395 (2006.01) 17.12.2014 Bulletin 2014/51 A61P 31/18 (2006.01) A61P 35/00 (2006.01) (21) Application number: 09745851.7 (86) International application number: PCT/EP2009/056026 (22) Date of filing: 18.05.2009 (87) International publication number: WO 2009/138519 (19.11.2009 Gazette 2009/47) (54) AMINO ACID SEQUENCES DIRECTED AGAINST CXCR4 AND OTHER GPCRs AND COMPOUNDS COMPRISING THE SAME GEGEN CXCR4 UND ANDERE GPCR GERICHTETE AMINOSÄURESEQUENZEN SOWIE VERBINDUNGEN DAMIT SÉQUENCES D’ACIDES AMINÉS DIRIGÉES CONTRE CXCR4 ET AUTRES GPCR ET COMPOSÉS RENFERMANT CES DERNIÈRES (84) Designated Contracting States: (74) Representative: Hoffmann Eitle AT BE BG CH CY CZ DE DK EE ES FI FR GB GR Patent- und Rechtsanwälte PartmbB HR HU IE IS IT LI LT LU LV MC MK MT NL NO PL Arabellastraße 30 PT RO SE SI SK TR 81925 München (DE) (30) Priority: 16.05.2008 US 53847 P (56) References cited: 02.10.2008 US 102142 P EP-A- 1 316 801 WO-A-99/50461 WO-A-03/050531 WO-A-03/066830 (43) Date of publication of application: WO-A-2006/089141 WO-A-2007/051063 23.02.2011 Bulletin 2011/08 • VADAY GAYLE G ET AL: "CXCR4 and CXCL12 (73) Proprietor: Ablynx N.V. (SDF-1) in prostate cancer: inhibitory effects of 9052 Ghent-Zwijnaarde (BE) human single chain Fv antibodies" CLINICAL CANCER RESEARCH, THE AMERICAN (72) Inventors: ASSOCIATION FOR CANCER RESEARCH, US, • BLANCHETOT, Christophe vol.10, no. -

Identification of Agonists for a Group of Human Odorant Receptors
ORIGINAL RESEARCH ARTICLE published: 03 March 2015 doi: 10.3389/fphar.2015.00035 Identification of agonists for a group of human odorant receptors Daniela C. Gonzalez-Kristeller 1,JoãoB.P.doNascimento1, Pedro A. F. Galante 2 and Bettina Malnic 1* 1 Department of Biochemistry, Institute of Chemistry, University of São Paulo, São Paulo, Brazil 2 Centro de Oncologia Molecular, Hospital Sírio-Libanês, São Paulo, Brazil Edited by: Olfaction plays a critical role in several aspects of the human life. Odorants are detected by Claudio M. Costa-Neto, University hundreds of odorant receptors (ORs) which belong to the superfamily of G protein-coupled of Sao Paulo, Brazil receptors. These receptors are expressed in the olfactory sensory neurons of the nose. Reviewed by: The information provided by the activation of different combinations of ORs in the nose Federico Monczor, Universidad de Buenos Aires, Argentina is transmitted to the brain, leading to odorant perception and emotional and behavioral Dietmar Krautwurst, Deutsche responses. There are ∼400 intact human ORs, and to date only a small percentage of Forschungsanstalt fuer these receptors (∼10%) have known agonists. The determination of the specificity of the Lebensmittelchemie Leibniz Institut, human ORs will contribute to a better understanding of how odorants are discriminated Germany by the olfactory system. In this work, we aimed to identify human specific ORs, that is, *Correspondence: Bettina Malnic, Department of ORs that are present in humans but absent from other species, and their corresponding Biochemistry, University of São agonists. To do this, we first selected 22 OR gene sequences from the human genome Paulo, Av. -

University of Copenhagen, Grønnegårdsvej 7, 1870 Frederiksberg, Denmark Cesses Covered Were Related to Each Other
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by Copenhagen University Research Information System Copy number variations and genome-wide associations reveal putative genes and metabolic pathways involved with feed conversion ratio in beef cattle Satana, Miguel Henrique de Almedia ; Junior, Gerson Antônio Oliveira ; Cesar, Aline Silva Mello; Freua, Mateus Castelani; Gomes, Rodrigo da Costra ; Silva, Saulo da Luz e ; Leme, Paulo Roberto; Fukumasu, Heidge; Carvalho, Minos Esperandio; Ventura, Ricardo Vierira; Coutinho, Luiz Lehmann; Kadarmideen, Haja; Ferraz, José Bento Sterman Published in: Journal of Applied Genetics DOI: 10.1007/s13353-016-0344-7 Publication date: 2016 Document license: Other Citation for published version (APA): Satana, M. H. D. A., Junior, G. A. O., Cesar, A. S. M., Freua, M. C., Gomes, R. D. C., Silva, S. D. L. E., ... Ferraz, J. B. S. (2016). Copy number variations and genome-wide associations reveal putative genes and metabolic pathways involved with feed conversion ratio in beef cattle. Journal of Applied Genetics, 57(4), 495- 504. https://doi.org/10.1007/s13353-016-0344-7 Download date: 08. apr.. 2020 J Appl Genetics DOI 10.1007/s13353-016-0344-7 ANIMAL GENETICS • ORIGINAL PAPER Copy number variations and genome-wide associations reveal putative genes and metabolic pathways involved with the feed conversion ratio in beef cattle Miguel Henrique de Almeida Santana1,2 & Gerson Antônio Oliveira Junior3 & Aline Silva Mello Cesar3 & Mateus Castelani Freua2 & Rodrigo da Costa Gomes4 & Saulo da Luz e Silva2 & Paulo Roberto Leme2 & Heidge Fukumasu2 & Minos Esperândio Carvalho2 & Ricardo Vieira Ventura2,5 & Luiz Lehmann Coutinho6 & Haja N. -
Explorations in Olfactory Receptor Structure and Function by Jianghai
Explorations in Olfactory Receptor Structure and Function by Jianghai Ho Department of Neurobiology Duke University Date:_______________________ Approved: ___________________________ Hiroaki Matsunami, Supervisor ___________________________ Jorg Grandl, Chair ___________________________ Marc Caron ___________________________ Sid Simon ___________________________ [Committee Member Name] Dissertation submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy in the Department of Neurobiology in the Graduate School of Duke University 2014 ABSTRACT Explorations in Olfactory Receptor Structure and Function by Jianghai Ho Department of Neurobiology Duke University Date:_______________________ Approved: ___________________________ Hiroaki Matsunami, Supervisor ___________________________ Jorg Grandl, Chair ___________________________ Marc Caron ___________________________ Sid Simon ___________________________ [Committee Member Name] An abstract of a dissertation submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy in the Department of Neurobiology in the Graduate School of Duke University 2014 Copyright by Jianghai Ho 2014 Abstract Olfaction is one of the most primitive of our senses, and the olfactory receptors that mediate this very important chemical sense comprise the largest family of genes in the mammalian genome. It is therefore surprising that we understand so little of how olfactory receptors work. In particular we have a poor idea of what chemicals are detected by most of the olfactory receptors in the genome, and for those receptors which we have paired with ligands, we know relatively little about how the structure of these ligands can either activate or inhibit the activation of these receptors. Furthermore the large repertoire of olfactory receptors, which belong to the G protein coupled receptor (GPCR) superfamily, can serve as a model to contribute to our broader understanding of GPCR-ligand binding, especially since GPCRs are important pharmaceutical targets. -

Novel Cell Lines and Methods
(19) TZZ¥ZZ_¥_T (11) EP 3 009 513 A1 (12) EUROPEAN PATENT APPLICATION (43) Date of publication: (51) Int Cl.: 20.04.2016 Bulletin 2016/16 C12N 15/85 (2006.01) C12N 15/67 (2006.01) C07K 14/435 (2006.01) (21) Application number: 15180871.4 (22) Date of filing: 02.02.2009 (84) Designated Contracting States: (72) Inventor: SHEKDAR, Kambiz AT BE BG CH CY CZ DE DK EE ES FI FR GB GR New York, NY 10010 (US) HR HU IE IS IT LI LT LU LV MC MK MT NL NO PL PT RO SE SI SK TR (74) Representative: Vossius & Partner Patentanwälte Rechtsanwälte mbB (30) Priority: 01.02.2008 US 63219 P Siebertstrasse 3 81675 München (DE) (62) Document number(s) of the earlier application(s) in accordance with Art. 76 EPC: Remarks: 09709529.3 / 2 245 171 •Claims filed after the date of filing of the application (Rule 68(4) EPC). (71) Applicant: Chromocell Corporation •This application was filed on 13-08-2015 as a North Brunswick, NJ 08902 (US) divisional application to the application mentioned under INID code 62. (54) NOVEL CELL LINES AND METHODS (57) The invention relates to novel cells and cell lines, and methods for making and using them. EP 3 009 513 A1 Printed by Jouve, 75001 PARIS (FR) EP 3 009 513 A1 Description Field of the Invention 5 [0001] The invention relates to novel cells and cell lines, and methods for making and using them. Background of the Invention [0002] Currently, the industry average failure rate for drug discovery programs in pharmaceutical companies is reported 10 to be approximately 98%. -

Supplementary Materialsupplementary Material
DOI:10.1071/AN20275_AC CSIRO 2021 Animal Production Science 2021, 61, 731–744 Genome-wide association study between copy number variation regions and carcass and meat quality traits in Nellore cattle Mariana Piatto Berton1,5, Marcos Vinícius de Antunes Lemos1, Tatiane Cristina Seleguim Chud2, Nedenia Bonvino Stafuzza1, Sabrina Kluska1, Sabrina Thaise Amorim1, Lucas Silva Ferlin Lopes1, Angélica Simone Cravo Pereira3, Derek Bickhart4, George Liu4, Lúcia Galvão de Albuquerque1 and Fernando Baldi1 1Departamento de Zootecnia, Faculdade de Ciências Agrárias e Veterinárias, Universidade Estadual Paulista, Via de acesso Prof. Paulo Donato Castellane, s/no, CEP 14884-900 Jaboticabal, SP, Brazil 2Centre for Genetic Improvement of Livestock, University of Guelph, Guelph, ON, Canada N1G 2W1 3 Faculdade de Zootecnia e Engenharia de Alimentos, Universidade de São Paulo, Rua Duque de Caxias Norte, 225, CEP 13635-900 Pirassununga, SP, Brazil 4 USDA-ARS, ANRI, Bovine Functional Genomics Laboratory, Beltsville, Maryland 20705, USA; 2Department of Animal and Avian Sciences, University of Maryland, College Park, Maryland 20742, USA 5Corresponding author. E-mail: [email protected] Table S1. Description of significant (P value<0.05) CNVRs associated with beef tenderness (BT) and candidate genes within the CNVRs. CNVR ID BTA Type* Start (bp) End (bp) Size (bp) Low** High** P value FDR CNVR_gain_3 1 Gain 1374155 3404571 2030417 27 45 0.026 0.36 CNVR_gain_644 2 Gain 81291862 83154452 1862591 32 15 0.015 0.40 CNVR_loss_936 5 Loss 22514133 22563988 49856 233