Pysal Documentation Release 1.14.3
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Sun Secure Global Desktop 4.5 Administration Guide
Sun Secure Global Desktop 4.5 Administration Guide Sun Microsystems, Inc. www.sun.com Part No. 820-6689-10 April 2009, Revision 01 Submit comments about this document at: http://docs.sun.com/app/docs/form/comments Copyright 2008-2009 Sun Microsystems, Inc., 4150 Network Circle, Santa Clara, California 95054, U.S.A. All rights reserved. Sun Microsystems, Inc. has intellectual property rights relating to technology that is described in this document. In particular, and without limitation, these intellectual property rights may include one or more of the U.S. patents listed at http://www.sun.com/patents and one or more additional patents or pending patent applications in the U.S. and in other countries. U.S. Government Rights - Commercial software. Government users are subject to the Sun Microsystems, Inc. standard license agreement and applicable provisions of the FAR and its supplements. This distribution may include materials developed by third parties. Parts of the product may be derived from Berkeley BSD systems, licensed from the University of California. UNIX is a registered trademark in the U.S. and in other countries, exclusively licensed through X/Open Company, Ltd. Sun, Sun Microsystems, the Sun logo, Solaris, OpenSolaris, Java, JavaScript, JDK, JavaServer Pages, JSP,JavaHelp, JavaBeans, JVM, JRE, Sun Ray, and StarOffice are trademarks or registered trademarks of Sun Microsystems, Inc. or its subsidiaries in the United States and other countries. All SPARC trademarks are used under license and are trademarks or registered trademarks of SPARC International, Inc. in the U.S. and in other countries. Products bearing SPARC trademarks are based upon an architecture developed by Sun Microsystems, Inc. -

Metadefender Core V4.17.3
MetaDefender Core v4.17.3 © 2020 OPSWAT, Inc. All rights reserved. OPSWAT®, MetadefenderTM and the OPSWAT logo are trademarks of OPSWAT, Inc. All other trademarks, trade names, service marks, service names, and images mentioned and/or used herein belong to their respective owners. Table of Contents About This Guide 13 Key Features of MetaDefender Core 14 1. Quick Start with MetaDefender Core 15 1.1. Installation 15 Operating system invariant initial steps 15 Basic setup 16 1.1.1. Configuration wizard 16 1.2. License Activation 21 1.3. Process Files with MetaDefender Core 21 2. Installing or Upgrading MetaDefender Core 22 2.1. Recommended System Configuration 22 Microsoft Windows Deployments 22 Unix Based Deployments 24 Data Retention 26 Custom Engines 27 Browser Requirements for the Metadefender Core Management Console 27 2.2. Installing MetaDefender 27 Installation 27 Installation notes 27 2.2.1. Installing Metadefender Core using command line 28 2.2.2. Installing Metadefender Core using the Install Wizard 31 2.3. Upgrading MetaDefender Core 31 Upgrading from MetaDefender Core 3.x 31 Upgrading from MetaDefender Core 4.x 31 2.4. MetaDefender Core Licensing 32 2.4.1. Activating Metadefender Licenses 32 2.4.2. Checking Your Metadefender Core License 37 2.5. Performance and Load Estimation 38 What to know before reading the results: Some factors that affect performance 38 How test results are calculated 39 Test Reports 39 Performance Report - Multi-Scanning On Linux 39 Performance Report - Multi-Scanning On Windows 43 2.6. Special installation options 46 Use RAMDISK for the tempdirectory 46 3. -

Oracle Secure Global Desktop 4.6 Administration Guide
® Oracle Secure Global Desktop Administration Guide for Version 4.6 Part No.: 821-1926-10 March 2011, Revision 02 Copyright © 2010, 2011, Oracle and/or its affiliates. All rights reserved. This software and related documentation are provided under a license agreement containing restrictions on use and disclosure and are protected by intellectual property laws. Except as expressly permitted in your license agreement or allowed by law, you may not use, copy, reproduce, translate, broadcast, modify, license, transmit, distribute, exhibit, perform, publish, or display any part, in any form, or by any means. Reverse engineering, disassembly, or decompilation of this software, unless required by law for interoperability, is prohibited. The information contained herein is subject to change without notice and is not warranted to be error-free. If you find any errors, please report them to us in writing. If this is software or related software documentation that is delivered to the U.S. Government or anyone licensing it on behalf of the U.S. Government, the following notice is applicable: U.S. GOVERNMENT RIGHTS Programs, software, databases, and related documentation and technical data delivered to U.S. Government customers are "commercial computer software" or "commercial technical data" pursuant to the applicable Federal Acquisition Regulation and agency-specific supplemental regulations. As such, the use, duplication, disclosure, modification, and adaptation shall be subject to the restrictions and license terms set forth in the applicable Government contract, and, to the extent applicable by the terms of the Government contract, the additional rights set forth in FAR 52.227-19, Commercial Computer Software License (December 2007). -

Oracle® Secure Global Desktop Platform Support and Release Notes for Version 4.62
Oracle® Secure Global Desktop Platform Support and Release Notes for Version 4.62 E23646-03 August 2012 Oracle® Secure Global Desktop: Platform Support and Release Notes for Version 4.62 Copyright © 2011, 2012, Oracle and/or its affiliates. All rights reserved. Oracle and Java are registered trademarks of Oracle and/or its affiliates. Other names may be trademarks of their respective owners. Intel and Intel Xeon are trademarks or registered trademarks of Intel Corporation. All SPARC trademarks are used under license and are trademarks or registered trademarks of SPARC International, Inc. AMD, Opteron, the AMD logo, and the AMD Opteron logo are trademarks or registered trademarks of Advanced Micro Devices. UNIX is a registered trademark of The Open Group. This software and related documentation are provided under a license agreement containing restrictions on use and disclosure and are protected by intellectual property laws. Except as expressly permitted in your license agreement or allowed by law, you may not use, copy, reproduce, translate, broadcast, modify, license, transmit, distribute, exhibit, perform, publish, or display any part, in any form, or by any means. Reverse engineering, disassembly, or decompilation of this software, unless required by law for interoperability, is prohibited. The information contained herein is subject to change without notice and is not warranted to be error-free. If you find any errors, please report them to us in writing. If this is software or related documentation that is delivered to the U.S. Government or anyone licensing it on behalf of the U.S. Government, the following notice is applicable: U.S. -

Basic UNIX 4: More on the GUI ● How X Works ● Window and Desktop Managers ● File Managers ● Common Tasks ● System Administration Tools
Basic UNIX 4: More on the GUI ● How X works ● Window and desktop managers ● File managers ● Common tasks ● System administration tools Iowa State University Information Technology Services Last update 1/29/2008 by jbalvanz How X works ● X server (X) – Provides tools for drawing graphics on a display – X applications send commands to the X server via TCP/IP ● X client – Machine running the software that wants to draw graphics – Usually the machine running the server, but doesn't have to be! How this looks Server Client Application package X Server TCP/IP TCP/IP Stack Stack Network Why is this good? ● Applications are independent of graphics hardware, window manager and even hardware platform ● Server and client do not have to be on the same machine; applications can be run remotely – Licensing considerations – Horsepower restrictions Running X applications remotely ● Connect to a remote machine using ssh with the -X option (not -x) ssh -X [email protected] ● Start the application sas Window managers ● Manages positioning of windows on the screen, virtual desktops, running applications; may include menus – AfterStep – Blackbox – Enlightenment Display Driver – FVWM Window Manager – IceWM – WindowMaker X Server – etc., etc., etc., etc. TCP/IP Stack Desktop Environments ● Window management + system utilities + standard applications + games + menuing system + ???????? – GNOME Desktop – K Desktop Environment (KDE) – Microsoft Windows What you'll have to get used to... ● Users have a choice of window/desktop manager (even in the stock RedHat install) ● Many desktop managers are themeable, i.e., FVWM may look completely different from FVWM on another machine depending on the choice of theme Why choice is good.. -

NOTICE: the Full Agenda Consists of Scanned Images of Only Those
Please click the links below to access the Council Agenda and Reports: 1. Council Agenda And Reports Documents: CAR 02-18-20.PDF MINS 02-03-20.PDF 02-18-20 2PM BRIEFING.PDF 2. City Council Agenda Documents: AG 02-18-20.PDF NOTICE: The Full Agenda consists of scanned images of only those reports and communications submitted to the City Clerk before the deadline established for such agenda and will not include any matter or item brought before Council for consideration at the meeting. The original documents are available for inspection in the Office of the City Clerk, Room 456 Noel C. Taylor Municipal Building, 215 Church Avenue, S. W., Roanoke, Virginia 24011. To receive the City Council agenda (without reports) automatically via e-mail, contact the Office of the City Clerk at [email protected] or (540) 853-2541. The records of City Council and City Clerk's Office will be maintained pursuant to Section 42.1-82 of the Code of Virginia (1950), as amended, and the Commonwealth of Virginia, Library of Virginia Records Management and Imaging Services Division, Records Retention and Disposition Schedules, for compliance with Guidelines provided by the Library of Virginia. ROANOKE CITY COUNCIL REGULAR SESSION FEBRUARY 18, 2020 2:00 P.M. CITY COUNCIL CHAMBER 215 CHURCH AVENUE, S. W. AGENDA 1. Call to Order--Roll Call. The Invocation will be delivered by The Reverend Eric Long, Pastor, St. John's Episcopal Church. The Pledge of Allegiance to the Flag of the United States of America will be led by Mayor Sherman P. -

UC Riverside UC Riverside Electronic Theses and Dissertations
UC Riverside UC Riverside Electronic Theses and Dissertations Title Extracting Actionable Information From Bug Reports Permalink https://escholarship.org/uc/item/4tf0c5x4 Author Zhou, Bo Publication Date 2016 Peer reviewed|Thesis/dissertation eScholarship.org Powered by the California Digital Library University of California UNIVERSITY OF CALIFORNIA RIVERSIDE Extracting Actionable Information From Bug Reports A Dissertation submitted in partial satisfaction of the requirements for the degree of Doctor of Philosophy in Computer Science by Bo Zhou December 2016 Dissertation Committee: Dr. Rajiv Gupta, Chairperson Dr. Iulian Neamtiu Dr. Zhiyun Qian Dr. Zhijia Zhao Copyright by Bo Zhou 2016 The Dissertation of Bo Zhou is approved: Committee Chairperson University of California, Riverside Acknowledgments This dissertation would not have been possible without all people who have helped me during my Ph.D. study and my life. Foremost, I would like to express my sincere gratitude to my advisor Dr. Iulian Neamtiu for the continuous support of my PhD study and research, for his patience, moti- vation, enthusiasm, and immense knowledge. I will never forget he is the person that went through my papers tens of times overnight with me for accuracy. His guidance helped me in all the time of research and writing of this dissertation. I could not have imagined having a better advisor and mentor for my PhD study. Thanks, Dr. Neamtiu! I am also deeply indebted to my co-advisor Dr. Rajiv Gupta for the collaboration over the years, guidance and help for making me not only a good researcher, but also a good person. His attitude towards research has influenced me immensely in the past and in the future. -
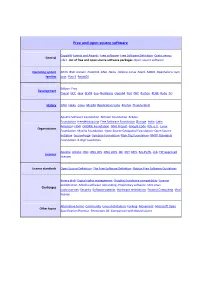
Free and Open Source Software
Free and open source software Copyleft ·Events and Awards ·Free software ·Free Software Definition ·Gratis versus General Libre ·List of free and open source software packages ·Open-source software Operating system AROS ·BSD ·Darwin ·FreeDOS ·GNU ·Haiku ·Inferno ·Linux ·Mach ·MINIX ·OpenSolaris ·Sym families bian ·Plan 9 ·ReactOS Eclipse ·Free Development Pascal ·GCC ·Java ·LLVM ·Lua ·NetBeans ·Open64 ·Perl ·PHP ·Python ·ROSE ·Ruby ·Tcl History GNU ·Haiku ·Linux ·Mozilla (Application Suite ·Firefox ·Thunderbird ) Apache Software Foundation ·Blender Foundation ·Eclipse Foundation ·freedesktop.org ·Free Software Foundation (Europe ·India ·Latin America ) ·FSMI ·GNOME Foundation ·GNU Project ·Google Code ·KDE e.V. ·Linux Organizations Foundation ·Mozilla Foundation ·Open Source Geospatial Foundation ·Open Source Initiative ·SourceForge ·Symbian Foundation ·Xiph.Org Foundation ·XMPP Standards Foundation ·X.Org Foundation Apache ·Artistic ·BSD ·GNU GPL ·GNU LGPL ·ISC ·MIT ·MPL ·Ms-PL/RL ·zlib ·FSF approved Licences licenses License standards Open Source Definition ·The Free Software Definition ·Debian Free Software Guidelines Binary blob ·Digital rights management ·Graphics hardware compatibility ·License proliferation ·Mozilla software rebranding ·Proprietary software ·SCO-Linux Challenges controversies ·Security ·Software patents ·Hardware restrictions ·Trusted Computing ·Viral license Alternative terms ·Community ·Linux distribution ·Forking ·Movement ·Microsoft Open Other topics Specification Promise ·Revolution OS ·Comparison with closed -

Southwestern M O N U M E N
10O3a SOUTHWESTERN MONUMENTS MONTHLY REPORT OCTOBER 19 3 9 UNITED STATES DEPARTMENT OF THE INTERIOR NATIONAL PARK SERVICE SOUTHWESTERN NATIONAL MONUMENTS QCTOBER.,1939 REPORT INDEX OPENING, by Superintendent Frank Pinkley 245 CONDENSED GENERAL REPJRT Travel 247 Activities of other agencies.250 100 Administrative... 248 400 Interpretation 251 200 Maintenance, New Con- 500 Use of monument facilities 252 struction, Improvements. .248 600 Protection 252 REPORTS FROM MEN IN THE FIELD Arches 253 Hovenweep-Yucca House 261 Aztec Ruins 260 Mobile Unit 280 Banaelier 289 Montezuma Castle 266 Bandelier CCC 294 Natural Bridges 295 Bandelier Ruins Stabilization.293 Navajo 277 Canyon de Chelly 284 Ortan Pipe Cactus 275 Capplin Moantain 269 Pipe Spring 256 Casa Grande 272 Saguart 270 Casa Grunue CCC 274 Sunset Crater 289 Chaco Canyon 279 Tontt, 258 Chaco Canyon CCC 281 Tumacacori 282 Chiricahua 258 Tuzigoot 262 Chiricahua CCC 259 Walnut Canyon 276 El Morro 295 Walnut Canyon CCC 277 Gran Quivira 264 White Sands 254 Wuputnl 286 HEADQUARTERS Branch of Accounting 304 Personnel notes 221 Branch of Historic Sites 299 October visitor statistics...219 Maintenance 304 Annual visitor statistics... .308 THE SUPPLEMENT Bird Nates from Banuelier, by G. G. Philp 314 Birds at Montezuma Castle, by Betty Jackson 312 Casta Grande Gleanings 311 Checa. List of Summer Flora at Chiricuhua, by Ora M. Clark 318 Chimings from the Chaco 314 Montezuma Muzings 313 Photographic Record Board, by Robert Lister 298 Ruminations, by the Boss 330 Siltin^s from the Sands 312 October, 1939 SOUTHWESTERN NATIONAL MONU m TS PERSOEKEL HEADQUARTERS ' ~ * " NATIONAL PARK SERVICE COOLIDGE, ARIZONA FRANK PINKLBY, SUPERINTENDENT Hugh M. -

Volume 131 December, 2017
Volume 131 December, 2017 HHaappppyy HHoolliiddaayyss!! In This Issue ... 3 From The Chief Editor's Desk ... 5 Paul's 2017 Holiday Gift Guide 8 Meemaw's 2017 Holiday Gift Guide 10 Screenshot Showcase 11 YouCanToo's 2017 Holiday Gift Guide 13 Screenshot Showcase 14 phorneker's 2017 Holiday Gift Guide 16 ms_meme's Nook: Bonnes Vacances C'est Si Bon 17 PCLinuxOS Recipe Corner: Beer Battered Grilled Cheese Sandwich 18 Google Cloud Print Capable Printer Using Raspberry Pi 21 Screenshot Showcase 22 Repo Review: Play It Slowly 22 Screenshot Showcase 23 Inkscape Tutorial: Creating A Pattern 25 GOG's Gems: Two Worlds - Epic Edition 28 Screenshot Showcase 29 Take Better Pictures With Your Smartphone 32 Is PCLinuxOS Really Safe In Today's Cyberworld? 35 Screenshot Showcase 36 ms_meme's Nook: Tex Is Good For You 37 PCLinuxOS Bonus Recipe Corner: Chicken Curry Salad Support PCLinuxOS! Get Your Official 38 PCLinuxOS Family Member Spotlight: Reziac PCLinuxOS 40 PCLinuxOS Puzzled Partitions Merchandise Today! 44 More Screenshot Showcase PCLinuxOS Magazine Page 2 From The Chief Editor's Desk ... It doesn’t seem like enough time has elapsed there in plenty of time, leaving work at 5:30 p.m. for the holidays to be coming around again, but Now the parking lot for this particular Wal-Mart they literally are just right around the corner. I is acres in size. She spent over 30 minutes know we’ve been busy buying the kids their trying to just find a parking spot, because every holiday gifts, as well as gifts for each other. We single one was filled. -

Opentext Exceed Turbox Release Notes
OpenText Exceed TurboX Release Notes Version 12 - 12.0.2 Product Released: April 2020 Contents 1 Introduction .................................................................................................................................... 9 1.1 Release Notes Revision History ................................................................................................ 9 1.2 Related documentation .............................................................................................................. 9 2 About OpenText Exceed TurboX ................................................................................................ 10 3 New features in Version 12.0.2 ................................................................................................... 10 3.1 Support for systemd ................................................................................................................ 10 3.2 Silent installation support for maximum number of sessions launched on a node ................. 10 3.3 Runtime removed from server patches ................................................................................... 10 3.4 New Connection Node state (Enabled, synchronizing) ........................................................... 11 3.5 ETX Server improvements ...................................................................................................... 11 3.5.1 Improved runtime deployment to connection nodes .................................................. 11 3.5.2 Improved login workflow .......................................................................................... -
Swm: an X Window Manager Shell 1. Introduction
swm: An X Window Manager Shell Thomas E. LaStrange Solbourne Computer Inc. 1900 Pike Road Longmont, CO 80501 [email protected] ABSTRACT swm is a policy-free, user con®gurable window manager client for the X Window System. Besides providing basic window manager functionality, swm introduces new features not found in existing window managers. First and foremost, swm has no default look-and-feel. Like the X Window system itself, swm does not dictate policy (look-and-feel); rather, it provides the mechanism for implementing window management policy. Users are not required to learn a new programming language to modify its behavior; instead, simple objects with associated actions determine swm's operation. Its major advantage over other window managers is a feature called the Virtual Desktop. The Virtual Desktop effectively makes the X root window larger than the physical limits of the display and can be panned in a number of ways, including scrollbars, a panner object, or window manager commands. Besides window management, swm also provides primitive session management. It can save a user's current window layout and restart those clients when X is restarted. swm can restart clients regardless of what toolkit they were built on or what remote host (if any) they were running on. All relevant client information is restored, including window position and size, icon position, and the state of the client. 1. Introduction Why another window manager? That is certainly a valid question given the number of window managers currently available for the X Window System1. Current window managers fall into two categories: easy to use but not very con®gurable, or very con®gurable but complicated to use.