Distribution of Nitrifying Bacteria Under Fluctuating Environmental Conditions
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Life Beyond Earth the Search for Habitable Worlds in the Universe
C:/ITOOLS/WMS/CUP-NEW/4181325/WORKINGFOLDER/COEN/9781107026179HTL.3D i [1–2] 27.6.2013 10:59AM Life Beyond Earth The Search for Habitable Worlds in the Universe With current missions to Mars and the Earth-like moon Titan, and many more missions planned, humankind stands on the verge of exciting progress and possible major discoveries in our quest for life in space. What is life and where can it exist? What searches are being made to identify conditions for life on other worlds? If extraterrestrial inhabited worlds are found, how can we explore them? Could humans survive beyond the Earth? In this book, two leading astrophysicists provide an engaging account of where we stand in our quest for habitable environments, in the Solar System and beyond. Starting from basic concepts, the narrative builds up scientifically, including more in-depth material as boxed additions to the main text. The authors recount fascinating recent discoveries, arising from space missions and from observations using ground-based telescopes, of possible life-related artefacts in Martian meteorites, of extrasolar planets, and of subsurface oceans on Europa, Titan and Enceladus. They also provide a forward look to exciting future missions, including the return to Venus, Mars and the Moon; further explorations of Pluto and Jupiter’s icy moons; and placing giant planet-seeking telescopes in orbit beyond Jupiter. showing how we approach the question of finding out whether the life that teems on our own planet is unique. This is an exciting, informative read for anyone interested in the search for habitable and inhabited planets, and makes an excellent primer for students keen to learn about astrobiology, habitability, planetary science and astronomy. -

Astrobiologia E a Busca Científica De Vida Fora Da Terra
Astronomia ao Meio Dia – IAG-USP 05/10/2017 Astrobiologia e a busca científica de vida fora da Terra Fabio Rodrigues Instituto de Química - USP Núcleo de Pesquisa em Astrobiologia - USP [email protected] Vida Extraterrestre • Prêmio Guzman (1900): 100.000 francos oferecido pela Académie des sciences (França) para quem conseguisse estabelecer comunicação extraterrestre. Curiosidade: Em seu texto, o prêmio excluia explícitamente a comunicação com Marte, pois neste planeta era óbvio que havia vida!! Evolução das observações de Marte! Christiaan Huygens, 1659 Telescopic view, 1960s Giovanni Schiaparelli, 1888 Hubble Space Telescope, 1997 Mars Global Surveyor, 2002 Furo com 1,6 cm de diâmetro! Especulativa Experimental / observacional Foto obtida pelo Mars Science Laboratoy (Curiosity) em 12 de Maio de 2014! O que é astrobiologia? Não é uma nova disciplina, mas um novo enfoque a antigos temas Origem e evolução da vida Distribição da vida, na Terra e fora dela Futuro da vida O que é astrobiologia? Não é uma nova disciplina, mas um novo enfoque a antigos temas Astrobiólogo?? - Físicos, químicos, biólogos, astrônomos, geólogos, cientistas planetários, filósofos, etc. - Interessados nos problemas e com o enfoque proposto pela astrobiologia! Big Bang Estrelas e supernovas Formação dos elementos Big Bang Estrelas e supernovas Moléculas Formação dos elementos (Astroquímica) + + - + + + 2 átomos: AlO, C2, CH, CH , CN, CN , CN , CO, CO , CS, FeO, H2, HCl, HCl , HO, OH , KCl, NH, + + N2, NO, NO , NaCl, MgH , O2, PO, SH, SO, SiC, SiN, SiO, TiO etc. + + 3 átomos: AlOH, C3, C2H, C2O, C2P, CO2, H3 , H2C, H2O, H2O , HO2, H2S, HCN, HNC, HCO, + + HCO+, HCP, HNC, HN2 , MgCN, NH2, N2H , N2O, NaOH, O3, SO2, SiC2, SiCN, TiO2 etc. -

Vida En Otros Planetas Eduardo Battaner Universidad De Granada
Vida en otros planetas Eduardo Battaner Universidad de Granada Estudiantes de Física y Electrónica EFE. Vida. Granada, 2011 ¿Vida en otros planetas? ● Sólo tenemos una prueba observacional. ● No sabemos cómo se produce la vida. Muchas gracias EFE. Vida. Granada, 2011 Hay planetas extrasolares ● Se han descubierto unos 500. ● Técnicas: Observación directa, efecto Doppler, astrometría, tránsitos. ● Corot, Kepler, Darwin (>2014) (espectrometría de atmósferas de planetas terrestres). ● Pero tienen vida? EFE. Vida. Granada, 2011 Misión Darwin ● ESA, 2014 ● 3 telescopios de 3m en interferometría, mili arcsec. ● Situado en L2 ● Infrarrojo. La estrella brilla sólo 1 millón de veces más. ● La estrella se anula interferométricamente. ● Espectro IR, oxígeno? Agua? ● La fotosíntesis produce oxígeno en La Tierra. ● Aunque puede producirse por fotodisociación del CO2. EFE. Vida. Granada, 2011 EFE. Vida. Granada, 2011 Titan ● Descubierto por Huygens (1655) ● Tiene atmósfera, Comás Solá (1908), por medio de ocultación. ● Diámetro 2000 km. Temperatura 90K ● Atmósfera de nitrógeno (94%) y metano. ● Orbitador Cassini, Sonda Huygens (2005) ● Montañas, ríos y mares (metano), islas ● Guijarros de hielo EFE. Vida. Granada, 2011 Titán Hicrocarburos procedentes de disociación de CH4 ¿Cuál es la fuente de CH4? Vientos muy rápidos como en Venus Tormentas: Sánchez Lavega, con descargas bruscas de metano. El CH4 se filtra por el suelo. EFE. Vida. Granada, 2011 EFE. Vida. Granada, 2011 ● ¿Fórmula de Drake ? ● ¿ªZona de habitabilidadº? ● ¿Oparin y Haldane? ● ¿Experimento de Miller y Urey? ● ¿Panspermia? ● Paradoja de Fermi. ● La ausencia de evidencia ¿es la evidencia de la ausencia? EFE. Vida. Granada, 2011 Gliese 581g ● Catálogo de Wilhelm Gliese (1957) < 25 pc ● Gliese 581 es de tipo M2 (-31ë , -12ë), 37 días. -
Goldilocks Planet
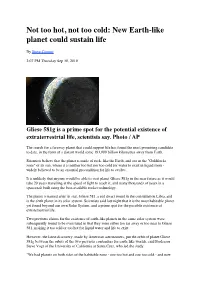
Not too hot, not too cold: New Earth-like planet could sustain life By Steve Connor 2:07 PM Thursday Sep 30, 2010 Gliese 581g is a prime spot for the potential existence of extraterrestrial life, scientists say. Photo / AP The search for a faraway planet that could support life has found the most promising candidate to date, in the form of a distant world some 193,000 billion kilometres away from Earth. Scientists believe that the planet is made of rock, like the Earth, and sits in the "Goldilocks zone" of its sun, where it is neither too hot nor too cold for water to exist in liquid form - widely believed to be an essential precondition for life to evolve. It is unlikely that anyone would be able to visit planet Gliese 581g in the near future as it would take 20 years travelling at the speed of light to reach it, and many thousands of years in a spacecraft built using the best-available rocket technology. The planet is named after its star, Gliese 581, a red dwarf found in the constellation Libra, and is the sixth planet in its solar system. Scientists said last night that it is the most habitable planet yet found beyond our own Solar System, and a prime spot for the possible existence of extraterrestrial life. Two previous claims for the existence of earth-like planets in the same solar system were subsequently found to be overstated in that they were either too far away or too near to Gliese 581, making it too cold or too hot for liquid water and life to exist. -

The Lick-Carnegie Exoplanet Survey: a 3.1 M Earth Planet in The
The Lick-Carnegie Exoplanet Survey: A 3.1 M⊕ Planet in the Habitable Zone of the Nearby M3V Star Gliese 581 Steven S. Vogt1, R. Paul Butler2, E. J. Rivera1, N. Haghighipour3, Gregory W. Henry4, and Michael H. Williamson4 Received ; accepted arXiv:1009.5733v1 [astro-ph.EP] 29 Sep 2010 1UCO/Lick Observatory, University of California, Santa Cruz, CA 95064 2Department of Terrestrial Magnetism, Carnegie Institution of Washington, 5241 Broad Branch Road, NW, Washington, DC 20015-1305 3Institute for Astronomy and NASA Astrobiology Institute, University of Hawaii-Manoa, Honolulu, HI 96822 4Tennessee State University, Center of Excellence in Information Systems, 3500 John A. Merritt Blvd., Box 9501, Nashville, TN. 37209-1561 –2– ABSTRACT We present 11 years of HIRES precision radial velocities (RV) of the nearby M3V star Gliese 581, combining our data set of 122 precision RVs with an ex- isting published 4.3-year set of 119 HARPS precision RVs. The velocity set now indicates 6 companions in Keplerian motion around this star. Differential photometry indicates a likely stellar rotation period of ∼ 94 days and reveals no significant periodic variability at any of the Keplerian periods, supporting planetary orbital motion as the cause of all the radial velocity variations. The combined data set strongly confirms the 5.37-day, 12.9-day, 3.15-day, and 67-day planets previously announced by Bonfils et al. (2005), Udry et al. (2007), and Mayor et al. (2009). The observations also indicate a 5th planet in the system, GJ 581f, a minimum-mass 7.0 M⊕ planet orbiting in a 0.758 AU orbit of period 433 days and a 6th planet, GJ 581g, a minimum-mass 3.1 M⊕ planet orbiting at 0.146 AU with a period of 36.6 days. -

Mathematics for Input Space Probes in the Atmosphere of Gliese 581D
Parana Journal of Science and Education, v.2, n.5, (6-13) July 14, 2016 PJSE, ISSN 2447-6153, c 2015-2016 https://sites.google.com/site/pjsciencea/ Mathematics for input space probes in the atmosphere of Gliese 581d R. Gobato1, A. Gobato2 and D. F. G. Fedrigo3 Abstract The work is a mathematical approach to the entry of an aerospace vehicle, as a probe or capsule in the atmosphere of the planet Gliese 581d, using data collected from the results of atmospheric models of the planet. GJ581d was the first planet candidate of a few Earth masses reported in the circum-stellar habitable zone of another star. It is located in the Gliese 581 star system, is a star red dwarf about 20 light years away from Earth in the constellation Libra. Its estimated mass is about a third of that of the Sun. It has been suggested that the recently discovered exoplanet GJ581d might be able to support liquid water due to its relatively low mass and orbital distance. However, GJ581d receives 35% less stellar energy than the planet Mars and is probably locked in tidal resonance, with extremely low insolation at the poles and possibly a permanent night side. The climate that demonstrate GJ581d will have a stable atmosphere and surface liquid water for a wide range of plausible cases, making it the first confirmed super-Earth (2-10 Earth masses) in the habitable zone. According to the general principle of relativity, “All systems of reference are equivalent with respect to the formulation of the fundamental laws of physics.” In this case all the equations studied apply to the exoplanet Gliese 581d. -

Open Research Online Oro.Open.Ac.Uk
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by Open Research Online Open Research Online The Open University’s repository of research publications and other research outputs Multiplication of microbes below 0.690 water activity: implications for terrestrial and extraterrestrial life Journal Item How to cite: Stevenson, Andrew; Burkhardt, Jürgen; Cockell, Charles S.; Cray, Jonathan A.; Dijksterhuis, Jan; Fox-Powell, Mark; Kee, Terence P.; Kminek, Gerhard; McGenity, Terry J.; Timmis, Kenneth N.; Timson, David J.; Voytek, Mary A.; Westall, Frances; Yakimov, Michail M. and Hallsworth, John E. (2015). Multiplication of microbes below 0.690 water activity: implications for terrestrial and extraterrestrial life. Environmental Microbiology, 17(2) pp. 257–277. For guidance on citations see FAQs. c 2014 Society for Applied Microbiology and John Wiley Sons Ltd Version: Version of Record Link(s) to article on publisher’s website: http://dx.doi.org/doi:10.1111/1462-2920.12598 Copyright and Moral Rights for the articles on this site are retained by the individual authors and/or other copyright owners. For more information on Open Research Online’s data policy on reuse of materials please consult the policies page. oro.open.ac.uk bs_bs_banner Environmental Microbiology (2015) 17(2), 257–277 doi:10.1111/1462-2920.12598 Minireview Multiplication of microbes below 0.690 water activity: implications for terrestrial and extraterrestrial life Andrew Stevenson,1 Jürgen Burkhardt,2 Summary Charles S. Cockell,3 Jonathan A. Cray,1 Since a key requirement of known life forms is avail- Jan Dijksterhuis,4 Mark Fox-Powell,3 Terence P. -

Habitability of the Goldilocks Planet Gliese 581G: Results from Geodynamic Models
Astronomy & Astrophysics manuscript no. final c ESO 2021 August 23, 2021 Habitability of the Goldilocks planet Gliese 581g: Results from geodynamic models (Research Note) W. von Bloh1, M. Cuntz2, S. Franck1, and C. Bounama1 1 Potsdam Institute for Climate Impact Research, P.O. Box 60 12 03, 14412 Potsdam, Germany e-mail: [email protected] 2 Department of Physics, University of Texas at Arlington, Box 19059, Arlington, TX 76019, USA Received ¡date¿ / Accepted ¡date¿ ABSTRACT Aims. In 2010, detailed observations have been published that seem to indicate another super-Earth planet in the system of Gliese 581 located in the midst of the stellar climatological habitable zone. The mass of the planet, known as Gl 581g, has been estimated to be between 3.1 and 4.3 M⊕. In this study, we investigate the habitability of Gl 581g based on a previously used concept that explores its long-term possibility of photosynthetic biomass production, which has already been used to gauge the principal possibility of life regarding the super-Earths Gl 581c and Gl 581d. Methods. A thermal evolution model for super-Earths is used to calculate the sources and sinks of atmospheric carbon dioxide. The habitable zone is determined by the limits of photosynthetic biological productivity on the planetary surface. Models with different ratios of land / ocean coverage are pursued. Results. The maximum time span for habitable conditions is attained for water worlds at a position of about 0.14 ± 0.015 AU, which deviates by just a few percent (depending on the adopted stellar luminosity) from the actual position of Gl 581g, an estimate that does however not reflect systematic uncertainties inherent in our model. -

Goldilocks’ Planet Has Potential for Life 10/08/2010
Newly Discovered ‘Goldilocks’ Planet Has Potential for Life 10/08/2010 http://www.pbs.org/newshour/extra/features/science/july-dec10/gliese_10-08.html Estimated Time: One 45-minute class period with possible extension Student Worksheet (reading comprehension and discussion questions without answers) PROCEDURE 1. WARM UP Use initiating questions to introduce the topic and find out how much your students know. 2. MAIN ACTIVITY Have students read NewsHour Extra's feature story and answer the reading comprehension and discussion questions on the student handout. 3. DISCUSSION Use discussion questions to encourage students to think about how the issues outlined in the story affect their lives and express and debate different opinions.] INITIATING QUESTIONS: 1. What is a planet? 2. How many planets are in the solar system? 3. How far is earth from the sun? READING COMPREHENSION: 1. What is the name of the new planet that American astronomers have discovered and how far away is it? Gliese 581g and 20 light years away 2. What are the names of the scientists who discovered the new planet and which institutions are they affiliated? Scientists Steven Vogt of the University of California and R. Paul Butler of the Carnegie Institution of Washington 3. How did scientists discover the planet? Vogt and Butler found the planet by studying the light from its parent star. Shifts in the light are caused by gravity from masses (planets) orbiting the star. 4. Why is Gliese 581g referred to as the “Goldilocks” planet? It is the perfect distance from its sun and therefore not too hot and not too cold -— just like porridge Goldilocks found at the home of the Three Bears. -

篇名:Life on Exoplanets?
投稿類別: 地球科學類 篇名: Life on Exoplanets? 作者: 葉庭妤。台北市立大安高工。綜高二忠 指導老師: 劉宗憲 Life on Exoplanets? I. Foreword 1. Motivation Galaxies, supernovas, extraterrestrials……how starry- eyed I used to be back in my childhood days. Exploring the unknown is extremely mystifying, yet it was only recently that I’ve learned the fact of existing astronomical objects outside the solar system. That’s why I’ve chosen this topic for my research essay. 2. Purpose of Research Many people are desperate to know if any Earth-like planet does appear so that when Earth faces an irrevocable catastrophe there would be a spare region for everyone to immigrate. Furthermore, the potential residents of the target place also prompt us to make more observations of other plausible forms of life and their development in reference to their survival. 3. Methodology As long as the Kepler mission remains active, new discoveries have sprung up like mushrooms each day. The frequently updated database of research essays therefore makes the best information in reach. II. Text Humans have long been pursuing another life-existing planet that is like our own, and ever since the first extrasolar planets (or ‘exoplanets’), the three planets orbiting around a pulsar (a dead star which emits wobbly light that makes it more attractive comparing to other stars) PSR 1257+1 were discovered twenty years ago, people started to have faith in finding candidates. 1. Types of exoplanets We can classify the planets, according to their characteristics, into eight types. Gas giants are the most commonly detected planets, being as big as Jupiter and Saturn, with a hydrogen and helium combination as a body and a metallic core. -

Twin Cities Creation Science Association
In the Beginning The Gauntlet is Thrown Down - Introduction Breaking Down the Equation . N = R* fp ne fl fi fc L Summary of Criticisms Conclusion Hand Outs Master Script Hand Out Life Outside Planet Earth 1961 How does the Drake Equation show that there is life on other planets in our Galaxy, or does it? N = the Number of civilizations in our galaxy with which radio- communication might be possible R* = the average Rate of star formation in our galaxy fp = the fraction of those stars that have planets ne = the average number of planets that can potentially support life per star that has planets (subscript e for ecoshell or earthlike) fl = the fraction of planets that could support life that actually develop life at some point fi = the fraction of planets with life that actually go on to develop intelligent life (in the form of civilizations) fc = the fraction of civilizations that develop a technology that releases detectable signs of their existence into space L = the Length of time for which such civilizations release detectable signals into space The Drake Equation . N = R* fp ne fl fi fc L Help! Can I exist? . N = R* fp ne fl fi fc L N is the number of detectable civilizations in our galaxy by a radio signal. Introduction Radio astronomer Frank Drake became the first person to start a systematic search for intelligent signals from the cosmos. Using the 25 meter dish of the National Radio Astronomy Observatory in Green Bank, West Virginia. Drake hosted a "search for extraterrestrial intelligence" (SETI) meeting on detecting their radio signals. -

Multiplication of Microbes Below 0.690 Water Activity: Implications for Terrestrial and Extraterrestrial Life
This is a repository copy of Multiplication of microbes below 0.690 water activity: implications for terrestrial and extraterrestrial life. White Rose Research Online URL for this paper: http://eprints.whiterose.ac.uk/121554/ Version: Accepted Version Article: Stevenson, A, Burkhardt, J, Cockell, CS et al. (12 more authors) (2015) Multiplication of microbes below 0.690 water activity: implications for terrestrial and extraterrestrial life. Environmental Microbiology, 17 (2). pp. 257-277. ISSN 1462-2912 https://doi.org/10.1111/1462-2920.12598 © 2014 Society for Applied Microbiology and John Wiley & Sons Ltd. This is the peer reviewed version of the following article: Stevenson, A., Burkhardt, J., Cockell, C. S., Cray, J. A., Dijksterhuis, J., Fox-Powell, M., Kee, T. P., Kminek, G., McGenity, T. J., Timmis, K. N., Timson, D. J., Voytek, M. A., Westall, F., Yakimov, M. M. and Hallsworth, J. E. (2015), Multiplication of microbes below 0.690 water activity: implications for terrestrial and extraterrestrial life. Environ Microbiol, 17: 257–277. doi:10.1111/1462-2920.12598, which has been published in final form at https://doi.org/10.1111/1462-2920.12598. This article may be used for non-commercial purposes in accordance with Wiley Terms and Conditions for Self-Archiving. Uploaded in accordance with the publisher's self-archiving policy. Reuse Unless indicated otherwise, fulltext items are protected by copyright with all rights reserved. The copyright exception in section 29 of the Copyright, Designs and Patents Act 1988 allows the making of a single copy solely for the purpose of non-commercial research or private study within the limits of fair dealing.