A New Methodology to Use Color Spectral Data for Taxonomic, Phylogenetic, and Biogeographic Studies
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Darwin Initiative Action Plan for the Coastal Biodiversity of Anegada, British Virgin Islands
Darwin Initiative Action Plan for the Coastal Biodiversity of Anegada, British Virgin Islands Darwin Anegada BAP 2006 Page We dedicate this document to the people of Anegada; the stewards of Anegada’s biodiversity and to Raymond Walker of the BVI National Parks Trust who tragically died after a very short illness during the course of this project. This report should be cited as: McGowan A., A.C.Broderick, C.Clubbe, S.Gore, B.J.Godley, M.Hamilton, B.Lettsome, J.Smith-Abbott, N.K.Woodfield. 2006. Darwin Initiative Action Plan for the Coastal Biodiversity of Anegada, British Virgin Islands. 13 pp. Available online at: http://www.seaturtle.org/mtrg/projects/anegada/ Darwin Anegada BAP 2006 Page 2 1. Introduction It well known that Anegada has globally important biodiversity. Indeed, biodiversity is the basis for most livelihoods; supporting fisheries and leading to the attractiveness that is such a draw to visitors. Over the last three years (2003-2006), a project was undertaken on Anegada with a wide range of activities focussing towards this Biodiversity Action Plan. From the outset it was known that the island hosts a globally important coral reef system, regionally significant populations of marine turtles, is of regional importance to birds and supports globally important endemic plants. The project arose following the encouragement of Anegada community members and subsequent extensive consultation between Dr. Godley (University of Exeter) and heads of BVI Conservation and Fisheries Department (CFD) and BVI National Parks Trust (NPT) who requested that funding be sourced for a project which: 1. Allowed the coastal biodiversity of Anegada to be assessed; 2. -
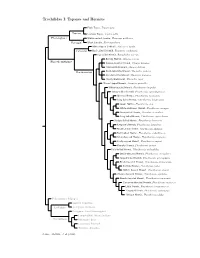
Topazes and Hermits
Trochilidae I: Topazes and Hermits Fiery Topaz, Topaza pyra Topazini Crimson Topaz, Topaza pella Florisuginae White-necked Jacobin, Florisuga mellivora Florisugini Black Jacobin, Florisuga fusca White-tipped Sicklebill, Eutoxeres aquila Eutoxerini Buff-tailed Sicklebill, Eutoxeres condamini Saw-billed Hermit, Ramphodon naevius Bronzy Hermit, Glaucis aeneus Phaethornithinae Rufous-breasted Hermit, Glaucis hirsutus ?Hook-billed Hermit, Glaucis dohrnii Threnetes ruckeri Phaethornithini Band-tailed Barbthroat, Pale-tailed Barbthroat, Threnetes leucurus ?Sooty Barbthroat, Threnetes niger ?Broad-tipped Hermit, Anopetia gounellei White-bearded Hermit, Phaethornis hispidus Tawny-bellied Hermit, Phaethornis syrmatophorus Mexican Hermit, Phaethornis mexicanus Long-billed Hermit, Phaethornis longirostris Green Hermit, Phaethornis guy White-whiskered Hermit, Phaethornis yaruqui Great-billed Hermit, Phaethornis malaris Long-tailed Hermit, Phaethornis superciliosus Straight-billed Hermit, Phaethornis bourcieri Koepcke’s Hermit, Phaethornis koepckeae Needle-billed Hermit, Phaethornis philippii Buff-bellied Hermit, Phaethornis subochraceus Scale-throated Hermit, Phaethornis eurynome Sooty-capped Hermit, Phaethornis augusti Planalto Hermit, Phaethornis pretrei Pale-bellied Hermit, Phaethornis anthophilus Stripe-throated Hermit, Phaethornis striigularis Gray-chinned Hermit, Phaethornis griseogularis Black-throated Hermit, Phaethornis atrimentalis Reddish Hermit, Phaethornis ruber ?White-browed Hermit, Phaethornis stuarti ?Dusky-throated Hermit, Phaethornis squalidus Streak-throated Hermit, Phaethornis rupurumii Cinnamon-throated Hermit, Phaethornis nattereri Little Hermit, Phaethornis longuemareus ?Tapajos Hermit, Phaethornis aethopygus ?Minute Hermit, Phaethornis idaliae Polytminae: Mangos Lesbiini: Coquettes Lesbiinae Coeligenini: Brilliants Patagonini: Giant Hummingbird Lampornithini: Mountain-Gems Tro chilinae Mellisugini: Bees Cynanthini: Emeralds Trochilini: Amazilias Source: McGuire et al. (2014).. -

List of and Notes on the Birds of the Iles Des Saintes, French West Indies
LIST OF AND NOTES ON THE BIRDS OF THE ILES DES SAINTES, FRENCH WEST INDIES CHARLES VAURIE LES Saintes,a small archipelagothat lies south of Guadeloupe,has seldolnbeen visited by ornithologists.No list of its birdsseems to have beenpublished, although, to besure, 25 specieshave been mentioned froin theseislands by Noble (1916), Bond (1936, 1956), Danforth(1939), or Pinchonand Bon Saint-Come(1951). Three of thesewere recorded on doubtfulevidence and, pendingconfirmation, should be deletedfrom the list. Nineteen of the remaining 22 and an additional five were observedby me on 2-5 July 1960,when Mrs. Vaurie and I visitedthe FrenchWest Indiesfor the AmericanMuseum of Natural History. The speciesmentioned by Noble and Danforth were incorporatedin their reportson the birdsof Guadeloupe,those cited by Bondor Pinchonand Bon Saint-Comein generalworks on the birds of the West Indies. The birds reportedso far are listedbelow with a few notes. I aln grateful to James Bond for his very helpful advice, Eugene Eisenmannfor readingthe manuscript,and to the Gendarmerieof Basse Terre and Terre-de-haut for arranging our visit as there is no hotel in Les Saintes. Les Saintesare of volcanicorigin and are separatedfrom the Basse Terre in Guadeloupeby a stormychannel seven miles wide. They con- sist of two relativelylarge islandscalled Terre-de-bas and Terre-de-haut, the only islandsinhabited, and of six small islandsor islets, some of which appear to be the sites of sea-bird colonies. One of these, called La Redonde,lies only 300 metersoff Terre-de-hautbut, unfortunately, is quite inaccessibleas it risesperpendicularly out of the sea to a height of 46 metersand is poundedby huge waves that rise to nearly half its height. -

Dominican Republic Endemics of Hispaniola II 1St February to 9Th February 2021 (9 Days)
Dominican Republic Endemics of Hispaniola II 1st February to 9th February 2021 (9 days) Palmchat by Adam Riley Although the Dominican Republic is perhaps best known for its luxurious beaches, outstanding food and vibrant culture, this island has much to offer both the avid birder and general naturalist alike. Because of the amazing biodiversity sustained on the island, Hispaniola ranks highest in the world as a priority for bird protection! This 8-day birding tour provides the perfect opportunity to encounter nearly all of the island’s 32 endemic bird species, plus other Greater Antillean specialities. We accomplish this by thoroughly exploring the island’s variety of habitats, from the evergreen and Pine forests of the Sierra de Bahoruco to the dry forests of the coast. Furthermore, our accommodation ranges from remote cabins deep in the forest to well-appointed hotels on the beach, each with its own unique local flair. Join us for this delightful tour to the most diverse island in the Caribbean! RBL Dominican Republic Itinerary 2 THE TOUR AT A GLANCE… THE ITINERARY Day 1 Arrival in Santo Domingo Day 2 Santo Domingo Botanical Gardens to Sabana del Mar (Paraiso Caño Hondo) Day 3 Paraiso Caño Hondo to Santo Domingo Day 4 Salinas de Bani to Pedernales Day 5 Cabo Rojo & Southern Sierra de Bahoruco Day 6 Cachote to Villa Barrancoli Day 7 Northern Sierra de Bahoruco Day 8 La Placa, Laguna Rincon to Santo Domingo Day 9 International Departures TOUR ROUTE MAP… RBL Dominican Republic Itinerary 3 THE TOUR IN DETAIL… Day 1: Arrival in Santo Domingo. -

1 AOS Classification Committee – North and Middle America Proposal Set 2020-A 4 September 2019 No. Page Title 01 02 Change Th
AOS Classification Committee – North and Middle America Proposal Set 2020-A 4 September 2019 No. Page Title 01 02 Change the English name of Olive Warbler Peucedramus taeniatus to Ocotero 02 05 Change the generic classification of the Trochilini (part 1) 03 11 Change the generic classification of the Trochilini (part 2) 04 18 Split Garnet-throated Hummingbird Lamprolaima rhami 05 22 Recognize Amazilia alfaroana as a species not of hybrid origin, thus moving it from Appendix 2 to the main list 06 26 Change the linear sequence of species in the genus Dendrortyx 07 28 Make two changes concerning Starnoenas cyanocephala: (a) assign it to the new monotypic subfamily Starnoenadinae, and (b) change the English name to Blue- headed Partridge-Dove 08 32 Recognize Mexican Duck Anas diazi as a species 09 36 Split Royal Tern Thalasseus maximus into two species 10 39 Recognize Great White Heron Ardea occidentalis as a species 11 41 Change the English name of Checker-throated Antwren Epinecrophylla fulviventris to Checker-throated Stipplethroat 12 42 Modify the linear sequence of species in the Phalacrocoracidae 13 49 Modify various linear sequences to reflect new phylogenetic data 1 2020-A-1 N&MA Classification Committee p. 532 Change the English name of Olive Warbler Peucedramus taeniatus to Ocotero Background: “Warbler” is perhaps the most widely used catch-all designation for passerines. Its use as a meaningful taxonomic indicator has been defunct for well over a century, as the “warblers” encompass hundreds of thin-billed, insectivorous passerines across more than a dozen families worldwide. This is not itself an issue, as many other passerine names (flycatcher, tanager, sparrow, etc.) share this common name “polyphyly”, and conventions or modifiers are widely used to designate and separate families that include multiple groups. -

Bird Checklist Guánica Biosphere Reserve Puerto Rico
United States Department of Agriculture BirD CheCklist Guánica Biosphere reserve Puerto rico Wayne J. Arendt, John Faaborg, Miguel Canals, and Jerry Bauer Forest Service Research & Development Southern Research Station Research Note SRS-23 The Authors: Wayne J. Arendt, International Institute of Tropical Forestry, U.S. Department of Agriculture Forest Service, Sabana Field Research Station, HC 2 Box 6205, Luquillo, PR 00773, USA; John Faaborg, Division of Biological Sciences, University of Missouri, Columbia, MO 65211-7400, USA; Miguel Canals, DRNA—Bosque de Guánica, P.O. Box 1185, Guánica, PR 00653-1185, USA; and Jerry Bauer, International Institute of Tropical Forestry, U.S. Department of Agriculture Forest Service, Río Piedras, PR 00926, USA. Cover Photos Large cover photograph by Jerry Bauer; small cover photographs by Mike Morel. Product Disclaimer The use of trade or firm names in this publication is for reader information and does not imply endorsement by the U.S. Department of Agriculture of any product or service. April 2015 Southern Research Station 200 W.T. Weaver Blvd. Asheville, NC 28804 www.srs.fs.usda.gov BirD CheCklist Guánica Biosphere reserve Puerto rico Wayne J. Arendt, John Faaborg, Miguel Canals, and Jerry Bauer ABSTRACt This research note compiles 43 years of research and monitoring data to produce the first comprehensive checklist of the dry forest avian community found within the Guánica Biosphere Reserve. We provide an overview of the reserve along with sighting locales, a list of 185 birds with their resident status and abundance, and a list of the available bird habitats. Photographs of habitats and some of the bird species are included. -

Flowers Visited by Hummingbirds in an Urban Cerrado Fragment, Mato Grosso Do Sul, Brazil
Biota Neotrop., vol. 13, no. 4 Flowers visited by hummingbirds in an urban Cerrado fragment, Mato Grosso do Sul, Brazil Waldemar Guimarães Barbosa-Filho1,2 & Andréa Cardoso de Araujo1 1Laboratório de Ecologia, Centro de Ciências Biológicas e da Saúde, Universidade Federal de Mato Grosso do Sul – UFMS, CP 549, CEP 79070-900, Campo Grande, MS, Brasil. http://www-nt.ufms.br/ 2Corresponding author: Waldemar Guimarães Barbosa-Filho, e-mail: [email protected] BARBOSA-FILHO, W.G. & ARAUJO, A.C. Flowers visited by hummingbirds in an urban Cerrado fragment, Mato Grosso do Sul, Brazil. Biota Neotrop. 13(4): http://www.biotaneotropica.org.br/v13n4/en/ abstract?article+bn00213042013 Abstract: Hummingbirds are the main vertebrate pollinators in the Neotropics, but little is known about the interactions between hummingbirds and flowers in areas of Cerrado. This paper aims to describe the interactions between flowering plants (ornithophilous and non-ornithophilous species) and hummingbirds in an urban Cerrado remnant. For this purpose, we investigated which plant species are visited by hummingbirds, which hummingbird species occur in the area, their visiting frequency and behavior, their role as legitimate or illegitimate visitors, as well as the number of agonistic interactions among these visitors. Sampling was conducted throughout 18 months along a track located in an urban fragment of Cerrado vegetation in Campo Grande, Mato Grosso do Sul, Brasil. We found 15 species of plants visited by seven species of hummingbirds. The main habit for ornithophilous species was herbaceous, with the predominance of Bromeliaceae; among non-ornithophilous most species were trees from the families Vochysiaceae and Malvaceae. -

The Journal of Caribbean Ornithology
THE J OURNAL OF CARIBBEAN ORNITHOLOGY SOCIETY FOR THE C ONSERVATION AND S TUDY OF C ARIBBEAN B IRDS S OCIEDAD PARA LA C ONSERVACIÓN Y E STUDIO DE LAS A VES C ARIBEÑAS ASSOCIATION POUR LA C ONSERVATION ET L’ E TUDE DES O ISEAUX DE LA C ARAÏBE 2005 Vol. 18, No. 1 (ISSN 1527-7151) Formerly EL P ITIRRE CONTENTS RECUPERACIÓN DE A VES M IGRATORIAS N EÁRTICAS DEL O RDEN A NSERIFORMES EN C UBA . Pedro Blanco y Bárbara Sánchez ………………....................................................................................................................................................... 1 INVENTARIO DE LA A VIFAUNA DE T OPES DE C OLLANTES , S ANCTI S PÍRITUS , C UBA . Bárbara Sánchez ……..................... 7 NUEVO R EGISTRO Y C OMENTARIOS A DICIONALES S OBRE LA A VOCETA ( RECURVIROSTRA AMERICANA ) EN C UBA . Omar Labrada, Pedro Blanco, Elizabet S. Delgado, y Jarreton P. Rivero............................................................................... 13 AVES DE C AYO C ARENAS , C IÉNAGA DE B IRAMA , C UBA . Omar Labrada y Gabriel Cisneros ……………........................ 16 FORAGING B EHAVIOR OF T WO T YRANT F LYCATCHERS IN T RINIDAD : THE G REAT K ISKADEE ( PITANGUS SULPHURATUS ) AND T ROPICAL K INGBIRD ( TYRANNUS MELANCHOLICUS ). Nadira Mathura, Shawn O´Garro, Diane Thompson, Floyd E. Hayes, and Urmila S. Nandy........................................................................................................................................ 18 APPARENT N ESTING OF S OUTHERN L APWING ON A RUBA . Steven G. Mlodinow................................................................ -

Ornithological Surveys in Serranía De Los Churumbelos, Southern Colombia
Ornithological surveys in Serranía de los Churumbelos, southern Colombia Paul G. W . Salaman, Thomas M. Donegan and Andrés M. Cuervo Cotinga 12 (1999): 29– 39 En el marco de dos expediciones biológicos y Anglo-Colombian conservation expeditions — ‘Co conservacionistas anglo-colombianas multi-taxa, s lombia ‘98’ and the ‘Colombian EBA Project’. Seven llevaron a cabo relevamientos de aves en lo Serranía study sites were investigated using non-systematic de los Churumbelos, Cauca, en julio-agosto 1988, y observations and standardised mist-netting tech julio 1999. Se estudiaron siete sitios enter en 350 y niques by the three authors, with Dan Davison and 2500 m, con 421 especes registrados. Presentamos Liliana Dávalos in 1998. Each study site was situ un resumen de los especes raros para cada sitio, ated along an altitudinal transect at c. 300- incluyendo los nuevos registros de distribución más m elevational steps, from 350–2500 m on the Ama significativos. Los resultados estabilicen firme lo zonian slope of the Serranía. Our principal aim was prioridad conservacionista de lo Serranía de los to allow comparisons to be made between sites and Churumbelos, y aluco nos encontramos trabajando with other biological groups (mammals, herptiles, junto a los autoridades ambientales locales con insects and plants), and, incorporating geographi cuiras a lo protección del marcizo. cal and anthropological information, to produce a conservation assessment of the region (full results M e th o d s in Salaman et al.4). A sizeable part of eastern During 14 July–17 August 1998 and 3–22 July 1999, Cauca — the Bota Caucana — including the 80-km- ornithological surveys were undertaken in Serranía long Serranía de los Churumbelos had never been de los Churumbelos, Department of Cauca, by two subject to faunal surveys. -

Colombia 1 000 Birds Mega Tour II 21St November to 19Th December 2014 (29 Days)
Colombia 1 000 Birds Mega Tour II 21st November to 19th December 2014 (29 days) Lance-tailed Manakin by Dennis Braddy Trip report compiled by Tour Leader: Rob Williams Trip Report - RBT Colombia Mega II 2014 2 An early start on day 1 saw us heading to Mundo Nuevo. Our first stop en route produced a flurry of birds including Northern Mountain Cacique, Golden-fronted Whitestart, Barred Becard, Mountain Elaenia and a Green-tailed Trainbearer feeding young at a nest. We continued up to the altitude where the endemic Flame-winged Parakeets breed and breakfasted while we awaited them. We were rewarded with great scope looks at this threatened species. The area also gave us a flurry of other birds including Scarlet-bellied Mountain Tanager, Rufous-breasted Chat- Tyrant, Pearled Treerunner and Smoke-coloured Pewee. We continued up to the edge of the paramo and birded a track inside Chingaza National Park. Activity was low but we persisted and were rewarded with a scattering of birds including Glossy, Masked and Bluish Flowerpiercers, Slaty Brush Finch, Glowing and Coppery-bellied Pufflegs, and Brown-backed Chat-Tyrant. The endemic Bronze-tailed Thornbill only gave frustrating brief flyby views. Great looks however were had of the endemic Pale-bellied Tapaculo, singing from surprisingly high up in a bush. The track back down gave us Rufous Wren, Superciliated and Black-capped Hemispingus and Tourmaline Sunangel. Further down the road a Buff- breasted Mountain Tanager and some Beryl-spangled Tanagers were found before we headed back to La Calera. After lunch in a local restaurant we headed to the Siecha gravel pits. -

Juan Cristóbal Gundlach's Collections of Puerto Rican Birds with Special
ZOBODAT - www.zobodat.at Zoologisch-Botanische Datenbank/Zoological-Botanical Database Digitale Literatur/Digital Literature Zeitschrift/Journal: Zoosystematics and Evolution Jahr/Year: 2015 Band/Volume: 91 Autor(en)/Author(s): Frahnert Sylke, Roman Rafela Aguilera, Eckhoff Pascal, Wiley James W. Artikel/Article: Juan Cristóbal Gundlach’s collections of Puerto Rican birds with special regard to types 177-189 Creative Commons Attribution 4.0 licence (CC-BY); original download https://pensoft.net/journals Zoosyst. Evol. 91 (2) 2015, 177–189 | DOI 10.3897/zse.91.5550 museum für naturkunde Juan Cristóbal Gundlach’s collections of Puerto Rican birds with special regard to types Sylke Frahnert1, Rafaela Aguilera Román2, Pascal Eckhoff1, James W. Wiley3 1 Museum für Naturkunde, Leibniz-Institut für Evolutions- und Biodiversitätsforschung, Invalidenstraße 43, D-10115 Berlin, Germany 2 Instituto de Ecología y Sistemática, La Habana, Cuba 3 PO Box 64, Marion Station, Maryland 21838-0064, USA http://zoobank.org/B4932E4E-5C52-427B-977F-83C42994BEB3 Corresponding author: Sylke Frahnert ([email protected]) Abstract Received 1 July 2015 The German naturalist Juan Cristóbal Gundlach (1810–1896) conducted, while a resident Accepted 3 August 2015 of Cuba, two expeditions to Puerto Rico in 1873 and 1875–6, where he explored the Published 3 September 2015 southwestern, western, and northeastern regions of this island. Gundlach made repre sentative collections of the island’s fauna, which formed the nucleus of the first natural Academic editor: history museums in Puerto Rico. When the natural history museums closed, only a few Peter Bartsch specimens were passed to other institutions, including foreign museums. None of Gund lach’s and few of his contemporaries’ specimens have survived in Puerto Rico. -

Volume 2. Animals
AC20 Doc. 8.5 Annex (English only/Seulement en anglais/Únicamente en inglés) REVIEW OF SIGNIFICANT TRADE ANALYSIS OF TRADE TRENDS WITH NOTES ON THE CONSERVATION STATUS OF SELECTED SPECIES Volume 2. Animals Prepared for the CITES Animals Committee, CITES Secretariat by the United Nations Environment Programme World Conservation Monitoring Centre JANUARY 2004 AC20 Doc. 8.5 – p. 3 Prepared and produced by: UNEP World Conservation Monitoring Centre, Cambridge, UK UNEP WORLD CONSERVATION MONITORING CENTRE (UNEP-WCMC) www.unep-wcmc.org The UNEP World Conservation Monitoring Centre is the biodiversity assessment and policy implementation arm of the United Nations Environment Programme, the world’s foremost intergovernmental environmental organisation. UNEP-WCMC aims to help decision-makers recognise the value of biodiversity to people everywhere, and to apply this knowledge to all that they do. The Centre’s challenge is to transform complex data into policy-relevant information, to build tools and systems for analysis and integration, and to support the needs of nations and the international community as they engage in joint programmes of action. UNEP-WCMC provides objective, scientifically rigorous products and services that include ecosystem assessments, support for implementation of environmental agreements, regional and global biodiversity information, research on threats and impacts, and development of future scenarios for the living world. Prepared for: The CITES Secretariat, Geneva A contribution to UNEP - The United Nations Environment Programme Printed by: UNEP World Conservation Monitoring Centre 219 Huntingdon Road, Cambridge CB3 0DL, UK © Copyright: UNEP World Conservation Monitoring Centre/CITES Secretariat The contents of this report do not necessarily reflect the views or policies of UNEP or contributory organisations.