Can Plastid Genome Sequencing Be Used for Species Identification in the Lauraceae?
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Cassytha Pubescens
Cassytha pubescens: Germination biology and interactions with native and introduced hosts Hong Tai (Steven), Tsang B.Sc. Hons (University of Adelaide) Thesis submitted for the degree of Master of Science School of Earth & Environmental Science University of Adelaide, Australia 03/05/2010 i Table of Contents Table of Contents ........................................................................................................... ii Abstract .......................................................................................................................... v Declaration ................................................................................................................... vii Acknowledgements .................................................................................................... viii Chapter. 1 Introduction .................................................................................................. 1 1.1 General Introduction ............................................................................................ 1 1.2 Literature Review ................................................................................................. 3 1.2.1 Characteristics of parasitic control agents .................................................... 3 1.2.2. Direct impacts on hosts ................................................................................ 7 1.2.3. Indirect impacts on hosts ............................................................................. 8 1.2.4. Summary ................................................................................................... -

The Litsea Genome and the Evolution of the Laurel Family
The Litsea genome and the evolution of the laurel family Chen et al 1 Supplementary Note 1. Sample preparation for Litsea cubeba genome sequencing For genome sequencing, we collected buds of L. cubeba. Genomic DNA was extracted using a modified cetyltrimethylammonium bromide (CTAB) protocol. For transcriptome analysis, we collected leaves, flowers, and roots from L. cubeba in Zhejiang Province, China, using a karyotype of 2n = 24 (Supplementary Figure 2a). Genome sizes can be determined from the total number of k-mers, divided by the peak value of the k-mer distribution1. To estimate the genome size of L. cubeba, we used a 350 bp pair-end library with 93.08 Gb high-quality reads to calculate the distribution of k-mer values, and found the main peak to be 54 (Supplementary Figure 2b). We estimated the L. cubeba genome size as 1370.14 Mbp, with a 1% heterozygosity rate and a 70.59% repeat sequence, based on an analysis of k-mer-numbers/depths. We used k-mer 41 to obtain a preliminary assembly of L. cubeba, with a scaffold N50 size of 776 bp and a corresponding contig N50 size of 591 bp. Supplementary Note 2. Whole genome duplication analysis in Laurales The KS peaks for WGDs in L. cubeba are both younger (smaller KS values) than the orthologous KS peak between L. cubeba and V. vinifera, implying that the two WGD events are specific to Magnoliids. To compare the WGD peaks of L. cubeba and the speciation events in the lineage of Magnoliids, we performed relative rate tests and corrected orthologous KS peaks between L. -

Fragrant Prickly-Apple Cereus Eriophorus Var
Fragrant Prickly-apple Cereus eriophorus var. fragrans (Small) L. Benson ereus eriophorus var. fragrans is a rare, slender, Federal Status: Endangered (Nov. 1, 1985) columnar cactus restricted to 11 small disjunct sites Critical Habitat: None Designated Cin eastern St. Lucie County. A 1996 survey Florida Status: Endangered documented the presence of 320 plants. Habitat loss and fragmentation remains a serious threat for plants on private Recovery Plan Status: Revision (May 18, 1999) lands. On public lands this species is protected from Geographic Coverage: Rangewide destruction, but in many areas it is experiencing a precipitous decline in abundance. This account represents a revision of the existing Figure 1. County distribution of the fragrant prickly-apple. recovery plan for the fragrant prickly-apple (FWS 1988). Description Cereus eriophorus var. fragrans is a solitary tree cactus that may have from one to eight, spiny, cane-like, stout, and succulent stems. The columnar stems are 2.5 to 5.0 cm in diameter, and have 10 or 12 ridges alternated with deep, sharp grooves (Benson 1982). Stems may be erect, or for longer stems, the plant may recline over neighboring vegetation. The branching can be extensive, and the roots of this cactus are coarse, fibrous, and shallow (Small 1920). The spine-bearing regions (areoles) are aligned along its ridges about 2 cm apart. Each areole bears 9 to 13 spines, which are mostly grayish and yellowish at the tip, with one spine longer (2 to 4 cm) than the rest. Cereus eriophorus var. fragrans has initial flower buds that are 1 cm long, white, and exceedingly hairy. -

Nesting of the Acrocephalus Warblers
Acta zoologica cracoviensia, 46(2): 97-195, Kraków, 30 June, 2003 Nesting of the Acrocephalus warblers Zygmunt BOCHEÑSKI and Piotr KUŒNIERCZYK Received: 14 Feb 2003 Accepted: 26 May 2003 BOCHEÑSKI Z., KUŒNIERCZYK P. 2003. Nesting of the Acrocephalus warblers. Acta zoo- logica cracoviensia, 46(2): 97-195. Abstract. The paper contains data concerning nest sites, material, construction, shape, and sizes in the majority of Acrocephalus species. The descriptions are based on field studies, museum specimens, and literature. The system proposed by CLEMENTS (2000) including 36 species in the genus Acrocephalus has been adopted. Similarities and differ- ences in nesting of 32 species and four subspecies are studied in the last chapter on the ba- sis of 38 characters assembled in Table XLI. They do not always reflect systematic relations of warblers within the genus Acrocephalus on the basis of molecular data. Key words: genus Acrocephalus, nest site, nest material, nest construction, nest shape, nest sizes. Z. BOCHEÑSKI (corresponding author), Institute of Systematics and Evolution of Ani- mals, P.Ac.Sc., 31-016 Kraków, S³awkowska 17, Poland. E-mail: [email protected] P. KUŒNIERCZYK, Ludwik Hirszfeld Institute of Immunology and Experimental Therapy, P. Ac.Sc., 53-114 Wroc³aw, Rudolfa Weigla 12, Poland. I. INTRODUCTION The genus Acrocephalus seems not to be defined unequivocally. From time to time it resembles a “witch’s sack”, to which various ornithologists place, according to their predilections, various species of Sylviinae, apart of a few, which are always in this genus. Those additional species are in- cluded in other genera, set up especially for them by other researchers . -

Nauru’S Biodiversity Strategy and Action Plan
FINAL DRAFT NAURU’S BIODIVERSITY STRATEGY AND ACTION PLAN Rehabilitate and Conserve Government of the Republic of Nauru 1 Nauru’s Biodiversity Strategy & Action Plan Prepared by: Dr Komeri Onorio and Tyrone Deiye for the National Environmental Coordinating Committee (NECC), Department of Commerce Industries and Environment Funded under: GEF/UNDP – Enabling Activities: The Convention on Biological Diversity 2 nauru vision for the future We are proceeding with the rehabilitation of our beloved island home in three overlapping and interlocking steps – physical, biological and cultural. Each step will be fully integrated with the other two so our process of future development rehabilitation and sustainable development will be interconnected. Physical rehabilitation must first deal with the land and water systems. The coral pinnacles will be dismantled, sawed and polished into building materials for homes and buildings. The coral pinnacles will also be crushed to make land fill, land will be graded, catchment areas and reservoirs built for storage of rainwater, and the freshwater underground lens are tapped for sustainable use. As part of the physical rehabilitation of Nauru island, topsoil, which we have stockpiled and carefully preserved, will be spread where it is needed for forests and fields, according to the land use plan that has been developed with maximum possible participation of all Nauruans. Biological rehabilitation will address the fields and forests, the coral reefs, and the surrounding seas. Areas of biological diversity will be established at strategically placed locations on the periphery of the island, and expanding gradually into the rainforest of tomorrow. Horticultural stations will be built at each of these strategic locations to nurse seedlings into trees, trees into forests, all according to the overall land use plan under development. -

The Stem Parasite Cassytha Filiformis a Botanical Relative of Avocado
California Avocado Society 1967 Yearbook 51: 159-160 THE STEM PARASITE CASSYTHA FILIFORMIS A BOTANICAL RELATIVE OF AVOCADO C. A. Schroeder University of California, Los Angeles The majority of genera and species which comprise the botanical family Lauraceae, of which the avocado is a commercially important member, are trees of variable size or woody shrubs or bushes. An unusual botanical relative in this interesting family of tropical and subtropical distribution is the "woevine," Cassytha filiformis. This species actually is a vine resembling the common parasitic dodder which is so prevalent in many parts of California and other subtropical and tropical areas of the world. While the habit is almost identical to Cuscuta, the floral structure is precisely like Cryptocarya, another botanical relative of the avocado, hence the genus is considered a member of the Lauraceae. The genus is chiefly Australian in more or less maritime climates. The "woevine" is cosmopolitan in the tropics where it sometimes becomes a pest of economic importance as it attaches itself to valuable orchard trees and ornamental plants with its tough threadlike extensive branchlets. It is a common stem parasite on Lantana Toddalia and certain other plants in South India (6). This plant is unique in the family in that it is a parasite. Because of its particular characteristics and its parasitic habit it is classified in a separate tribe. Cassytheae, within the family Lauraceae and is represented by the single genus Cassytha (2) in which 18 species have been described (3). The genus derives its name, Cassytha, from the Greek name for Cuscuta. Probably the most important and prominent species is C. -

Rapid Biodiversity Assessment of REPUBLIC of NAURU
RAPID BIODIVERSITY ASSESSMENT OF REPUBLIC OF NAURU JUNE 2013 NAOERO GO T D'S W I LL FIRS SPREP Library/IRC Cataloguing-in-Publication Data McKenna, Sheila A, Butler, David J and Wheatley, Amanda. Rapid biodiversity assessment of Republic of Nauru / Sheila A. McKeena … [et al.] – Apia, Samoa : SPREP, 2015. 240 p. cm. ISBN: 978-982-04-0516-5 (print) 978-982-04-0515-8 (ecopy) 1. Biodiversity conservation – Nauru. 2. Biodiversity – Assessment – Nauru. 3. Natural resources conservation areas - Nauru. I. McKeena, Sheila A. II. Butler, David J. III. Wheatley, Amanda. IV. Pacific Regional Environment Programme (SPREP) V. Title. 333.959685 © SPREP 2015 All rights for commercial / for profit reproduction or translation, in any form, reserved. SPREP authorises the partial reproduction or translation of this material for scientific, educational or research purposes, provided that SPREP and the source document are properly acknowledged. Permission to reproduce the document and / or translate in whole, in any form, whether for commercial / for profit or non-profit purposes, must be requested in writing. Secretariat of the Pacific Regional Environment Programme P.O. Box 240, Apia, Samoa. Telephone: + 685 21929, Fax: + 685 20231 www.sprep.org The Pacific environment, sustaining our livelihoods and natural heritage in harmony with our cultures. RAPID BIODIVERSITY ASSESSMENT OF REPUBLIC OF NAURU SHEILA A. MCKENNA, DAVID J. BUTLER, AND AmANDA WHEATLEY (EDITORS) NAOERO GO T D'S W I LL FIRS CONTENTS Organisational Profiles 4 Authors and Participants 6 Acknowledgements -
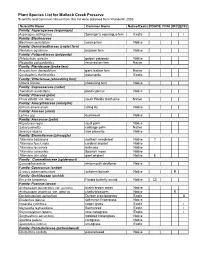
Plant Species List for Mullock Creek Preserve Scientific and Common Names from This List Were Obtained from Wunderlin 2003
Plant Species List for Mullock Creek Preserve Scientific and Common names from this list were obtained from Wunderlin 2003. Scientific Name Common Name Native/Exotic FDACS FNAI IRC EPPC Family: Asparagaceae (asparagus) Asparagus aethiopicus Sprenger's asparagus-fern Exotic I Family: Blechnaceae Blechnum serrulatum swamp fern Native Family: Dennstaedtiaceae (cuplet fern) Pteridium aquilinum bracken fern Native Family: Polypodiaceae (polypody) Phlebodium aureum golden polypody Native Pleopeltis polypodioides resurrection fern Native Family: Pteridaceae (brake fern) Acrostichum danaeifolium giant leather fern Native Ceratopteris thalictroides watersprite Exotic Family: Vittariaceae (shoestring fern) Vittaria lineata shoestring fern Native Family: Cupressaceae (cedar) Taxodium ascendens pond cypress Native Family: Pinaceae (pine) Pinus elliottii var. densa south Florida slash pine Native Family: Amaryllidaceae (amaryllis) Crinum americanum string lily Native Family: Araceae (arum) Lemna spp duckweed Native Family: Arecaceae (palm) Roystonea regia royal palm Native Sabal palmetto cabbage palm Native Serenoa repens saw palmetto Native Family: Bromeliaceae (pineapple) Tillandsia balbisiana northern needleleaf Native T Tillandsia fasciculata cardinal airplant Native Tillandsia recurvata ballmoss Native Tillandsia usneoides Spanish moss Native Tillandsia utriculata giant airplant Native E Family: Commelinaceae (spiderwort) Commelina erecta whitemouth dayflower Native Family: Cyperaceae (sedge) Scirpus tabernaemontani softstem bulrush Native R Family: -

A Taxonomic Revision of Cassytha (Lauraceae)
J. Adelaide Bot. Gard. 3(3): 187-262(1981) A TAXONOMIC REVISION OF CASSYTHA(LAURACEAE) IN AUSTRALIA J. Z. Weber State Herbarium, Botanic Gardens, North Terrace, Adelaide,South Australia 5000 Abstract The taxonomy of the genus Cassytha L. (Lauraceae) in Australia isrevised: in Australia there are 14 species, 3 varieties and 5 forms, all of them endemic except C. filiformis,C. capillaris and C. pubescens. The following new taxa are described: Cassytha aurea, C. pedicellosa, C.peninsularis, C. rufa, C. aurea var. candida, C. aurea var. hirta, C. peninsularis var.flindersii and C. glabella f. bicallosa. New combinations are: Cassytha glabella f. casuarinae (Nees) J.Z. Weber (C.casuarinae Nees), C. glabella f. dispar (Schltdl.) J.Z. Weber (C. dispar Schltd1.). C.racemosa f. muelleri (Meisn.) J.Z. Weber (C. muelleri Meisn.), C. racemosa f. pilosa (Benth.) J.Z. Weber (C.racemosa var. pilosa Benth.). An introduction includes discussion of morphology and includes the firstchromosome count for a species of Cassytha (C. pubescens: n= 24). The affinities of the species are discussed, their distribution described,a key to the species, varieties and forms provided and a revised description of each taxon is supplemented withdrawings, distributions maps and comment on possible evolutionary trends. Contents Introduction 187 Brief History of Taxonomy of Cassytha 188 Taxonomic Criteria 192 Biology 199 Distribution 200 Origins 201 Taxonomic Concepts 202 Taxonomic Treatment 202 Acknowledgements 256 References 260 Introduction Species of Cassytha share a remarkable similarity ofmorphology and habit with Cuscuta (Convolvulaceae), but considerable differences inflower, fruit, physiological and anatomical features show that this similarity is onlya case of convergent evolution to a parasitic habit. -

Host Specificity of Cassytha Filiformis and C. Pergracilis (Lauraceae) In
Bull. Natl. Mus. Nat. Sci., Ser. B, 38(2), pp. 47–53, May 22, 2012 Host Specificity of Cassytha filiformis and C. pergracilis (Lauraceae) in the Ryukyu Archipelago Goro Kokubugata1,* and Masatsugu Yokota2 1 Department of Botany, National Museum of Nature and Science, Tsukuba, Ibaraki 305–0005, Japan 2 Laboratory of Ecology and Systematics, Faculty of Science, University of the Ryukyus, Nishihara, Okinawa 903–0213, Japan * E-mail: [email protected] (Received 16 February 2012; accepted 28 March 2012) Abstract Host plants of two parasitic Cassytha species in the Ryukyu Archipelago, namely the pantropic Cassytha filiformis and the Ryukyu endemic C. pergracilis, were investigated in the present study. Twenty-six vascular species of pteridophytes and spermatophytes were recognized as host plants of C. filiformis. On the other hand, two monocotyledonous species Aristida takeoi (Poaceae) and Rhynchospora rubra (Cyperaceae), specifically occurring in a certain special envi- ronment in the Ryukyus, were recognized as host plants of C. pergracilis. Rhynchospora rubra is reported for the first time as a host plant of C. pergracilis. The present study implies that rarity of the Zwischenmoor vegetation in the Ryukyus and high habitat specificity of the two host species could be the cause of the narrow-distribution range of C. pergracilis. Key words : Cassytha, host specificity, parasitic plant, Ryukyus. Introduction duce a few short lateral roots using nutrients stored in the cotyledons. Once the first hausto- Parasitic plants derive some or all of their rium forms, the radicles disappear and the plant energy from other plants, and about 4100 species becomes independent of soil contact (Heide-Jor- of parasitic flowering plants in 270 genera are gensen, 2008). -

Phylogenetic Origins of Parasitic Plants Daniel L. Nickrent Chapter 3, Pp. 29-56 In: J. A. López-Sáez, P. Catalán and L
Phylogenetic Origins of Parasitic Plants Daniel L. Nickrent Chapter 3, pp. 29-56 in: J. A. López-Sáez, P. Catalán and L. Sáez [eds.], Parasitic Plants of the Iberian Peninsula and Balearic Islands This text first written in English (as appears here) was translated into Spanish and appeared in the book whose Spanish citation is: Nickrent, D. L. 2002. Orígenes filogenéticos de las plantas parásitas. Capitulo 3, pp. 29-56 In J. A. López-Sáez, P. Catalán and L. Sáez [eds.], Plantas Parásitas de la Península Ibérica e Islas Baleares. Mundi-Prensa Libros, S. A., Madrid. Throughout its history, the field of plant systematics has undergone changes in response to the advent of new philosophical ideas, types of data, and methods of analysis. It is no exaggeration to say that the past decade has witnessed a virtual revolution in phylogenetic investigation, owing mainly to the application of molecular methodologies and advancements in data analysis techniques. These powerful approaches have provided a source of data, independent of morphology, that can be used to address long-standing questions in angiosperm evolution. These new methods have been applied to systematic and phylogenetic questions among parasitic plants (Nickrent et al. 1998), but have often raised as many new questions as they have solved, in part due to the amazingly complex nature of the genetic systems present in these organisms. The goal of this chapter is to provide a general synopsis of the current state of understanding of parasitic plant phylogeny. To place in context results concerning the parasites, it is necessary to first examine general features of angiosperm phylogeny. -

Wild Plants for Survival in South Florida
MORTON: SURVIVAL PLANTS 313 WILD PLANTS FOR SURVIVAL IN SOUTH FLORIDA Julia F. Morton is the article, "Those Bounteous Florida Keys," Director, Morton Collectanea, written for the June, 1954, issue of Everglades Natural History by John D. Dickson III, based University of Miami on his personal experimentation while sta tioned for 14 months on Big Pine Key. Coral Gables Between 1944 and 1946, Dr. Richard A. The American Indians and early settlers, as Howard, now Director of the Arnold Arbore well as many more recent and contemporary tum of Harvard University, was Chief of the woodsmen, campers, adventurers and natural Survival Section, Air Force Tactical Air Center ists, have provided ample precedent for the at Orlando, Fla. In writing survival manuals utilization of our diverse and abundant indige and in teaching survival and rescue techniques nous plant life to augment the diet as well as for the U. S. Air Force, Dr. Howard (then meet many other human needs. Fortunately, Captain Howard) and his "students" collected the varied food and other uses of the principal native and introduced plant materials in wild plants of the United States as a whole southern Florida, worked out ways of ex have been ably recorded in a number of well- tracting and preparing their edible portions, known works, chiefly Yanovsky's Food Plants and ate such dishes during week-long survival of the North American Indians, Saunders' Use training periods. The food habits of the ful Wild Plants of the United States and Seminoles and more recent "natives" were Canada, Medsger's Edible Wild Plants, the examined and compared with the habits of "Edible Plants" chapter in Kephart's classic people of other tropical countries in the use of Camping and Woodcraft, the two editions of such widespread plants as palms, papayas, Edible Wild Plants of Eastern North America grasses, beach and marsh plants, and even sea by Fernald and Kinsey, the last edition revised weeds.