Development of a Binding Assay Between the HIV-1 Envelope
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Impact of Natural HIV-1 Nef Alleles and Polymorphisms on SERINC3/5 Downregulation
Impact of natural HIV-1 Nef alleles and polymorphisms on SERINC3/5 downregulation by Steven W. Jin B.Sc., Simon Fraser University, 2016 Thesis Submitted in Partial Fulfillment of the Requirements for the Degree of Master of Science in the Master of Science Program Faculty of Health Sciences © Steven W. Jin 2019 SIMON FRASER UNIVERSITY Spring 2019 Copyright in this work rests with the author. Please ensure that any reproduction or re-use is done in accordance with the relevant national copyright legislation. Approval Name: Steven W. Jin Degree: Master of Science Title: Impact of natural HIV-1 Nef alleles and polymorphisms on SERINC3/5 downregulation Examining Committee: Chair: Kanna Hayashi Assistant Professor Mark Brockman Senior Supervisor Associate Professor Masahiro Niikura Supervisor Associate Professor Ralph Pantophlet Supervisor Associate Professor Lisa Craig Examiner Professor Department of Molecular Biology and Biochemistry Date Defended/Approved: April 25, 2019 ii Ethics Statement iii Abstract HIV-1 Nef is a multifunctional accessory protein required for efficient viral pathogenesis. It was recently identified that the serine incorporators (SERINC) 3 and 5 are host restriction factors that decrease the infectivity of HIV-1 when incorporated into newly formed virions. However, Nef counteracts these effects by downregulating SERINC from the cell surface. Currently, there lacks a comprehensive study investigating the impact of primary Nef alleles on SERINC downregulation, as most studies to date utilize lab- adapted or reference HIV strains. In this thesis, I characterized and compared SERINC downregulation from >400 Nef alleles isolated from patients with distinct clinical outcomes and subtypes. I found that primary Nef alleles displayed a dynamic range of SERINC downregulation abilities, thus allowing naturally-occurring polymorphisms that modulate this activity to be identified. -

Download Article PDF/Slides
Kan Lu, PharmD New Antiretrovirals for Based on a presentation at prn by Roy M. Gulick, md, mph the Treatment of HIV: Kan Lu, PharmD | Drug Development Fellow University of North Carolina School of Pharmacy Chapel Hill, North Carolina The View in 2006 Roy M. Gulick, md, mph Reprinted from The prn Notebook® | october 2006 | Dr. James F. Braun, Editor-in-Chief Director, Cornell Clinical Trials Unit | Associate Professor of Medicine, Meri D. Pozo, PhD, Managing Editor. Published in New York City by the Physicians’ Research Network, Inc.® Weill Medical College of Cornell University | New York, New York John Graham Brown, Executive Director. For further information and other articles available online, visit http://www.prn.org | All rights reserved. ©october 2006 substantial progress continues to be made in the arena of cokinetics and a long extracellular half-life of approximately 10 hours antiretroviral drug development. prn is again proud to present its annual (Zhu, 2003). During apricitabine’s development, a serious drug interac- review of the experimental agents to watch for in the coming months and tion with lamivudine (Epivir) was noted. Although the plasma years. This year’s review is based on a lecture by Dr. Roy M. Gulick, a long- concentrations of apricitabine were unaffected by coadministration of time friend of prn, and no stranger to the antiretroviral development lamivudine, the intracellular concentrations of apricitabine were reduced pipeline. by approximately sixfold. Additionally, the 50% inhibitory concentration To date, twenty-two antiretrovirals have been approved by the Food (ic50) of apricitabine against hiv with the M184V mutation was increased and Drug Administration (fda) for the treatment of hiv infection. -

MS Ritgerð Aðalbjörg Aðalbjörnsdóttir
The Vif protein of maedi-visna virus Protein interaction and new roles Aðalbjörg Aðalbjörnsdóttir Thesis for the degree of Master of Science University of Iceland Faculty of medicine School of Health Sciences Vif prótein mæði-visnuveiru Prótein tengsl og ný hlutverk Aðalbjörg Aðalbjörnsdóttir Ritgerð til meistaragráðu í Líf og læknavísindum Umsjónarkennari: Valgerður Andrésdóttir Meistaranámsnefnd: Stefán Ragnar Jónsson og Ólafur S. Andrésson Læknadeild Heilbrigðisvísindasvið Háskóla Íslands Júní 2016 The Vif protein of maedi-visna virus Protein interaction and new roles Aðalbjörg Aðalbjörnsdóttir Thesis for the degree of Master of Science Supervisor: Valgerður Andrésdóttir Masters committee: Stefán Ragnar Jónsson and Ólafur S. Andrésson Faculty of Medicine School of Health Sciences June 2016 Ritgerð þessi er til meistaragráðu í Líf og læknavísindum og er óheimilt að afrita ritgerðina á nokkurn hátt nema með leyfi rétthafa. © Aðalbjörg Aðalbjörnsdóttir 2016 Prentun: Háskólaprent Reykjavík, Ísland 2016 Ágrip Mæði-visnuveira (MVV) er lentiveira af ættkvísl retróveira. Hún veldur hæggengri lungnabólgu (mæði) og heilabólgu (visnu) í kindum. Aðalmarkfrumur veirunnar eru mónocytar/makrófagar. Veiran er náskyld HIV og hefur verið notuð sem módel fyrir HIV sýkingar. Stöðug vopnakapphlaup milli veira og fruma hafa leitt af sér fjölda sértækra aðferða í vörnum hýsilsfrumu gegn veirusýkingum. Fruman hefur þróað með sér innrænar varnir gegn ýmsum sýkingum. Þessar varnir geta verið mjög sérhæfðar og tjáning þeirra spilar stórt hlutverk í hvaða frumur er hægt að sýkja og hverjar ekki. Dæmi um slíkan frumubundinn þátt eru APOBEC3 próteinin. APOBEC3 próteinin eru fjölskylda cytósín deaminasa sem geta hindrað retróveirur og retróstökkla. Þetta gera þau með því að afaminera cýtósín í úrasil í einþátta DNA á meðan á víxlritun stendur og valda þar með G-A stökkbreytingum í forveirunni. -

718 HIV Disorders of the Brain; Pathology and Pathogenesis Luis
[Frontiers in Bioscience 11, 718-732, January 1, 2006] HIV disorders of the brain; pathology and pathogenesis Luis Del Valle and Sergio Piña-Oviedo Center for Neurovirology and Cancer Biology, Laboratory of Neuropathology and Molecular Pathology, Temple University, 1900 North 12th Street, Suite 240, Philadelphia, Pennsylvania 19122 USA TABLE OF CONTENTS 1. Abstract 2. AIDS-Encephalopathy 2.1. Definition 2.2. HIV-1 Structure 2.3. Histopathology 2.4. Clinical Manifestations 2.5. Physiopathology 3. Progressive Multifocal Leukoencephalopathy 3.1. Definition 3.2. JC Virus Biological Considerations 3.3. JC Virus Structure 3.4. Histopathology 3.5. Clinical Manifestations 3.6. Physiopathology 4. Cryptococcosis 5.1. Definition 5.2. Cryptococcus neoformans Structure 5.3. Histopathology 5.4. Clinical Manifestations 5.5. Physiopathology 5. Toxoplasmosis 5.1. Definition 5.2. Toxoplasma godii Structure 5.3. Histopathology 5.4. Clinical Manifestations 5.5. Physiopathology 6. Primary CNS Lymphomas 6.1. Definition 6.2. Histopathology 6.3. Clinical Manifestations 6.4. Physiopathology 7. Acknowledgments 8. References 1. ABSTRACT Infection with HIV-1 has spread exponentially in still present in approximately 70 to 90% of patients and recent years to reach alarming proportions. It is estimated can be the result of HIV itself or of opportunistic than more than 33 million adults and 1.3 million children infections. Here we briefly review the pathology and are infected worldwide. Approximately 16,000 new cases pathophysiology of AIDS-Encephalopathy, of some of are diagnosed every day and almost 3 million people die the significant opportunistic infections affecting the every year from AIDS, making it the fourth leading brain in the context of AIDS, including Progressive cause of death in the world. -

UC Merced UC Merced Undergraduate Research Journal
UC Merced UC Merced Undergraduate Research Journal Title Antiviral Drugs Targeting Host Proteins an Efficient Strategy Permalink https://escholarship.org/uc/item/66f5b4m0 Journal UC Merced Undergraduate Research Journal, 9(2) Author Karmonphet, Arrada Publication Date 2017 DOI 10.5070/M492034789 Undergraduate eScholarship.org Powered by the California Digital Library University of California Antiviral Drugs Targeting Host Proteins an Efficient Strategy Arrada Karmonphet University of California, Merced Keywords: Proteins, Viruses, Drugs 1 Abstract Viruses have the ability to spread rapidly because the proteins and enzymes from the host cell help in the development of viruses. Although there are many vaccines that can prevent some viruses from infecting the body, the antiviral drugs today have not been effective in combating viruses from the start of spreading. This is due to the fact that the processes inside a virus are still being studied. However, host proteins proved to be valuable factors responsible for viral replication and spreading. It was found that certain functions such as capsid formation of the virus utilized a biochemical pathway that involved host proteins and some proteins of the host cell were evolutionarily conserved. When the important host proteins were altered, or removed the viruses weren’t able to replicate as effectively. It was concluded that targeting the host proteins had a significant effect in viral replication. This approach can stop viral replication from the start, create less viral resistance, and help find new antiviral drugs that work for many different types of viruses. This review will analyze five research articles about protein interactions in viruses and how monitoring the proteins and biochemical pathways can lead to the discovery of druggable targets during development. -

Tim Horn Deputy Executive Director, HIV & HCV Programs Treatment
Tim Horn Deputy Executive Director, HIV & HCV Programs Treatment Action Group NASTAD Prevention and Care Technical Assistance Meeting Washington, DC July 19, 2017 ▪ Pipeline is robust! ▪ Several drugs, coformulations, and biologics in late-stage development and Phase I trials ▪ Trends are clear ▪ Maximizing safety and efficacy of three-drug regimens ▪ Validating two-drug regimens as durable maintenance therapy and, potentially, for PLWHIV starting treatment for the first time ▪ Advancing long-acting and extended release products ▪ Development new drugs and biologics for multi-drug/class-resistant HIV ▪ Cost considerations in high- and middle-income countries Compound Class/Type Company Status DRUGS Isentress HD INSTI Merck FDA Approved 5/30/17 Bictegravir plus TAF/FTC INSTI plus NtRTI & NRTI Gilead Phase III; mid-2018 approval Doravirine (plus TDF/3TC) NNRTI (plus NtRTI & NRTI) Merck Phase III; mid-2018 approval Darunavir plus cobicistat, PI plus PK booster, NtRTI & Janssen Phase III; mid-2018 approval TAF & FTC NRTI Dolutegravir plus rilpivirine INSTI plus NNRTI ViiV/Jansse Phase III; early 2018 approval n Dolutegravir plus lamivudine INSTI plus NRTI ViiV Phase III; late 2018 approval BIOLOGICS Ibalizumab Entry Inhibitor TaiMed Phase III; late 2017 or early Biologics 2018 Compound Class/Type Company Status DRUGS LA Cabotegravir + INSTI + NNRTI ViiV/ Phase III LA Rilpivirine Janssen Albuvirtide Fusion Inhibitor Frontier Phase III; China is primary Biologics launch target Fostemsavir CD4 attachment inhibitor ViiV Phase III Elsulfavirine -

Assessment of the Interaction Between the Human
VARIATIONS IN THE V3 CROWN OF HIV-1 ENVELOPE IMPACT AFFINITY FOR CCR5 AND AFFECT ENTRY AND REPLICATIVE FITNESS By MICHAEL ANDREW LOBRITZ Submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy Dissertation advisor: Eric J. Arts, Ph.D. Department of Molecular Biology and Microbiology CASE WESTERN RESERVE UNIVERSITY August 2007 CASE WESTERN RESERVE UNIVERSITY SCHOOL OF GRADUATE STUDIES We hereby approve the dissertation of ______________________________________________________ candidate for the Ph.D. degree *. (signed)_______________________________________________ (chair of the committee) ________________________________________________ ________________________________________________ ________________________________________________ ________________________________________________ ________________________________________________ (date) _______________________ *We also certify that written approval has been obtained for any proprietary material contained therein. Table of Contents Chapter 1: Introduction........................................................................................................................16 1.A. HIV and AIDS..............................................................................................17 1.B. Retroviruses: Structure, Organization, and Replication...............................20 1.B.1. HIV-1 Genome...............................................................................20 1.B.2. HIV-1 Particle................................................................................23 -

Autophagy and Mammalian Viruses: Roles in Immune Response, Viral Replication, and Beyond
Zurich Open Repository and Archive University of Zurich Main Library Strickhofstrasse 39 CH-8057 Zurich www.zora.uzh.ch Year: 2016 Autophagy and mammalian viruses: roles in immune response, viral replication, and beyond Paul, P ; Münz, C Abstract: Autophagy is an important cellular catabolic process conserved from yeast to man. Double- membrane vesicles deliver their cargo to the lysosome for degradation. Hence, autophagy is one of the key mechanisms mammalian cells deploy to rid themselves of intracellular pathogens including viruses. How- ever, autophagy serves many more functions during viral infection. First, it regulates the immune response through selective degradation of immune components, thus preventing possibly harmful overactivation and inflammation. Additionally, it delivers virus-derived antigens to antigen-loading compartments for presentation to T lymphocytes. Second, it might take an active part in the viral life cycle by, eg, facili- tating its release from cells. Lastly, in the constant arms race between host and virus, autophagy is often hijacked by viruses and manipulated to their own advantage. In this review, we will highlight key steps during viral infection in which autophagy plays a role. We have selected some exemplary viruses and will describe the molecular mechanisms behind their intricate relationship with the autophagic machinery, a result of host–pathogen coevolution. DOI: https://doi.org/10.1016/bs.aivir.2016.02.002 Posted at the Zurich Open Repository and Archive, University of Zurich ZORA URL: https://doi.org/10.5167/uzh-131236 Book Section Accepted Version The following work is licensed under a Creative Commons: Attribution-NonCommercial-NoDerivatives 4.0 International (CC BY-NC-ND 4.0) License. -

Role of Reverse Transcriptase and APOBEC3G in Survival of Human
& My gy co lo lo ro g i y V Soni et al., Virol Mycol 2013, 3:1 Virology & Mycology DOI: 10.4172/2161-0517.1000125 ISSN: 2161-0517 Review Article Open Access Role of Reverse Transcriptase and APOBEC3G in Survival of Human Immune Deficiency Virus -1 Genome Raj Kumar Soni*, Amol Kanampalliwar and Archana Tiwari School of Biotechnology, Rajiv Gandhi Proudyogiki Vishwavidyalaya (State Technological University), Madhya Pradesh, India Abstract Development of an effective vaccine against HIV-1 is a major challenge for scientists at present. Rapid mutation and replication of the virus in patients contribute to the evolution of the virus, which makes it unconquerable. Hence a deep understanding of critical elements related to HIV-1 is necessary. Errors introduced during DNA synthesis by reverse transcriptase are the primary source of genetic variation within retroviral populations. Numerous current studies have shown that apolipo protein B mRNA-editing enzyme-catalytic polypeptide-like 3G (APOBEC3G) proteins mediated sub- lethal mutagenesis of HIV-1 proviral DNA contributes in viral fitness by accelerating human immunodeficiency virus-1 evolution. This results in the loss of the immunity and development of resistance against anti-viral drugs. This review focuses on the latest biological, biochemical, and structural studies in an attempt to discuss current ideas related to mutations initiated by reverse transcriptase and APOBEC3G. It also describes their effect on immunological diversity and retroviral restriction, and their overall effect on the viral genome respectively. A new procedure for eradication of HIV-1 has also been proposed based on the previous studies and proven facts. Keywords: HIV-1; Reverse transcriptase; Recombination; defined. -

Reviewing HIV-1 Gag Mutations in Protease Inhibitors Resistance: Insights for Possible Novel Gag Inhibitor Designs
molecules Review Reviewing HIV-1 Gag Mutations in Protease Inhibitors Resistance: Insights for Possible Novel Gag Inhibitor Designs Chinh Tran-To Su 1, Darius Wen-Shuo Koh 1 and Samuel Ken-En Gan 1,2,* 1 Antibody & Product Development Lab, Bioinformatics Institute, A*STAR, Singapore 138671, Singapore 2 p53 Laboratory, A*STAR, Singapore 138648, Singapore * Correspondence: [email protected]; Tel.: +65-6478-8417; Fax: +65-6478-9047 Academic Editor: Keykavous Parang Received: 23 August 2019; Accepted: 4 September 2019; Published: 6 September 2019 Abstract: HIV protease inhibitors against the viral protease are often hampered by drug resistance mutations in protease and in the viral substrate Gag. To overcome this drug resistance and inhibit viral maturation, targeting Gag alongside protease rather than targeting protease alone may be more efficient. In order to successfully inhibit Gag, understanding of its drug resistance mutations and the elicited structural changes on protease binding needs to be investigated. While mutations on Gag have already been mapped to protease inhibitor resistance, there remain many mutations, particularly the non-cleavage mutations, that are not characterized. Through structural studies to unravel how Gag mutations contributes to protease drug resistance synergistically, it is thus possible to glean insights to design novel Gag inhibitors. In this review, we discuss the structural role of both novel and previously reported Gag mutations in PI resistance, and how new Gag inhibitors can be designed. Keywords: HIV-1 Gag; Gag inhibitors; protease; protease inhibitors; drug resistance mutations; drug design 1. Introduction Many anti-HIV drugs interfere directly with the viral life cycle by targeting key viral enzymes [1], e.g., reverse transcriptase inhibitors [2,3], integrase inhibitors [4,5], and protease inhibitors [6,7]. -

To Reduce HIV-1 Infectivity Facilitates Its Encapsidation Into the Virions GANP Interacts with APOBEC3G
GANP Interacts with APOBEC3G and Facilitates Its Encapsidation into the Virions To Reduce HIV-1 Infectivity This information is current as Kazuhiko Maeda, Sarah Ameen Almofty, Shailendra Kumar of September 23, 2021. Singh, Mohammed Mansour Abbas Eid, Mayuko Shimoda, Terumasa Ikeda, Atsushi Koito, Phuong Pham, Myron F. Goodman and Nobuo Sakaguchi J Immunol 2013; 191:6030-6039; Prepublished online 6 November 2013; Downloaded from doi: 10.4049/jimmunol.1302057 http://www.jimmunol.org/content/191/12/6030 Supplementary http://www.jimmunol.org/content/suppl/2013/11/06/jimmunol.130205 http://www.jimmunol.org/ Material 7.DC1 References This article cites 52 articles, 20 of which you can access for free at: http://www.jimmunol.org/content/191/12/6030.full#ref-list-1 Why The JI? Submit online. by guest on September 23, 2021 • Rapid Reviews! 30 days* from submission to initial decision • No Triage! Every submission reviewed by practicing scientists • Fast Publication! 4 weeks from acceptance to publication *average Subscription Information about subscribing to The Journal of Immunology is online at: http://jimmunol.org/subscription Permissions Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html Email Alerts Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts The Journal of Immunology is published twice each month by The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2013 by The American Association of Immunologists, Inc. All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. The Journal of Immunology GANP Interacts with APOBEC3G and Facilitates Its Encapsidation into the Virions To Reduce HIV-1 Infectivity Kazuhiko Maeda,*,1 Sarah Ameen Almofty,*,1 Shailendra Kumar Singh,* Mohammed Mansour Abbas Eid,* Mayuko Shimoda,* Terumasa Ikeda,†,2 Atsushi Koito,† Phuong Pham,‡ Myron F. -
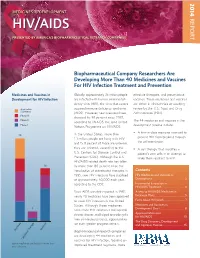
14-258 Phrma HIV/AIDS2014 0819.Indd
2014 MEDICINES IN DEVELOPMENT REPORT HIV/AIDS PRESENTED BY AMERICA’S BIOPHARMACEUTICAL RESEARCH COMPANIES Biopharmaceutical Company Researchers Are Developing More Than 40 Medicines and Vaccines For HIV Infection Treatment and Prevention Medicines and Vaccines in Globally, approximately 35 million people effective therapies, and preventative Development for HIV Infection are infected with human immunodefi - vaccines. These medicines and vaccines ciency virus (HIV), the virus that causes are either in clinical trials or awaiting Application acquired immune defi ciency syndrome review by the U.S. Food and Drug Submitted (AIDS). However, new infections have Administration (FDA). Phase III dropped by 38 percent since 2001, Phase II The 44 medicines and vaccines in the according to UNAIDS, the Joint United Phase I development pipeline include: Nations Programme on HIV/AIDS. • A fi rst-in-class medicine intended to In the United States, more than 25 prevent HIV from breaking through 1.1 million people are living with HIV the cell membrane. and 15.8 percent of those are unaware they are infected, according to the • A cell therapy that modifi es a U.S. Centers for Disease Control and patient’s own cells in an attempt to Prevention (CDC). Although the U.S. make them resistant to HIV. HIV/AIDS-related death rate has fallen 16 by more than 80 percent since the introduction of antiretroviral therapies in Contents 1995, new HIV infections have stabilized HIV Medicines and Vaccines in at approximately 50,000 each year, Development ......................................2 according to the CDC. Incremental Innovation in HIV/AIDS Treatment .......................... 4 Since AIDS was fi rst reported in 1981, Access to HIV/AIDS Medicines in nearly 40 medicines have been approved Exchange Plans ...................................5 to treat HIV infection in the United Facts About HIV/AIDS ........................7 States.