Nucleotide Metabolism
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Nucleotide Degradation
Nucleotide Degradation Nucleotide Degradation The Digestion Pathway • Ingestion of food always includes nucleic acids. • As you know from BI 421, the low pH of the stomach does not affect the polymer. • In the duodenum, zymogens are converted to nucleases and the nucleotides are converted to nucleosides by non-specific phosphatases or nucleotidases. nucleases • Only the non-ionic nucleosides are taken & phospho- diesterases up in the villi of the small intestine. Duodenum Non-specific phosphatases • In the cell, the first step is the release of nucleosides) the ribose sugar, most effectively done by a non-specific nucleoside phosphorylase to give ribose 1-phosphate (Rib1P) and the free bases. • Most ingested nucleic acids are degraded to Rib1P, purines, and pyrimidines. 1 Nucleotide Degradation: Overview Fate of Nucleic Acids: Once broken down to the nitrogenous bases they are either: Nucleotides 1. Salvaged for recycling into new nucleic acids (most cells; from internal, Pi not ingested, nucleic Nucleosides acids). Purine Nucleoside Pi aD-Rib 1-P (or Rib) 2. Oxidized (primarily in the Phosphorylase & intestine and liver) by first aD-dRib 1-P (or dRib) converting to nucleosides, Bases then to –Uric Acid (purines) –Acetyl-CoA & Purine & Pyrimidine Oxidation succinyl-CoA Salvage Pathway (pyrimidines) The Salvage Pathways are in competition with the de novo biosynthetic pathways, and are both ANABOLISM Nucleotide Degradation Catabolism of Purines Nucleotides: Nucleosides: Bases: 1. Dephosphorylation (via 5’-nucleotidase) 2. Deamination and hydrolysis of ribose lead to production of xanthine. 3. Hypoxanthine and xanthine are then oxidized into uric acid by xanthine oxidase. Spiders and other arachnids lack xanthine oxidase. -

35 Disorders of Purine and Pyrimidine Metabolism
35 Disorders of Purine and Pyrimidine Metabolism Georges van den Berghe, M.- Françoise Vincent, Sandrine Marie 35.1 Inborn Errors of Purine Metabolism – 435 35.1.1 Phosphoribosyl Pyrophosphate Synthetase Superactivity – 435 35.1.2 Adenylosuccinase Deficiency – 436 35.1.3 AICA-Ribosiduria – 437 35.1.4 Muscle AMP Deaminase Deficiency – 437 35.1.5 Adenosine Deaminase Deficiency – 438 35.1.6 Adenosine Deaminase Superactivity – 439 35.1.7 Purine Nucleoside Phosphorylase Deficiency – 440 35.1.8 Xanthine Oxidase Deficiency – 440 35.1.9 Hypoxanthine-Guanine Phosphoribosyltransferase Deficiency – 441 35.1.10 Adenine Phosphoribosyltransferase Deficiency – 442 35.1.11 Deoxyguanosine Kinase Deficiency – 442 35.2 Inborn Errors of Pyrimidine Metabolism – 445 35.2.1 UMP Synthase Deficiency (Hereditary Orotic Aciduria) – 445 35.2.2 Dihydropyrimidine Dehydrogenase Deficiency – 445 35.2.3 Dihydropyrimidinase Deficiency – 446 35.2.4 Ureidopropionase Deficiency – 446 35.2.5 Pyrimidine 5’-Nucleotidase Deficiency – 446 35.2.6 Cytosolic 5’-Nucleotidase Superactivity – 447 35.2.7 Thymidine Phosphorylase Deficiency – 447 35.2.8 Thymidine Kinase Deficiency – 447 References – 447 434 Chapter 35 · Disorders of Purine and Pyrimidine Metabolism Purine Metabolism Purine nucleotides are essential cellular constituents 4 The catabolic pathway starts from GMP, IMP and which intervene in energy transfer, metabolic regula- AMP, and produces uric acid, a poorly soluble tion, and synthesis of DNA and RNA. Purine metabo- compound, which tends to crystallize once its lism can be divided into three pathways: plasma concentration surpasses 6.5–7 mg/dl (0.38– 4 The biosynthetic pathway, often termed de novo, 0.47 mmol/l). starts with the formation of phosphoribosyl pyro- 4 The salvage pathway utilizes the purine bases, gua- phosphate (PRPP) and leads to the synthesis of nine, hypoxanthine and adenine, which are pro- inosine monophosphate (IMP). -

Xanthine Oxidase Assay (XO) Cat
Xanthine Oxidase Assay (XO) Cat. No. 8458 100 Tests in 96-well plate Introduction Xanthine Oxidase (XO) located predominantly in the liver and intestine in mammalian tissues and catalyzes the hydroxylation of hypoxanthine to xanthine and then to uric acid and hydrogen peroxide. XO activity is normally very low in blood and liver injury can result in the release of XO into blood. XO activity or expression can be up-regulated in gout and cardiovascular disease. This colorimetric assay is based on XO-catalyzed oxidation of xanthine, in which the formed hydrogen peroxide is catalyzed by peroxidase and reacts with 4-aminoantipyrine to form the product dye. The color intensity of the reaction product at 550nm is directly proportional to XO activity in the sample. Kit Components Cat. No. # of vials Reagent Quantity Storage 8458a 1 Assay buffer 10 mL 4°C 8458b 1 Xanthine Oxidase standard 0.2 mL -20°C 8458c 1 Xanthine 2.0 mL -20°C 8458d 1 Substrate mix 1.6 mL -20°C 8458e 1 Enzyme mix 0.1 mL -20°C Product Use The Xanthine Oxidase Assay kit measures the xanthine oxidase level of different types of samples, such as serum, plasma, tissues. This product is for research purposes only and not for use in animals, humans, or diagnostic procedures. Quality Control Serially diluted xanthine oxidase solutions with concentrations ranging from 7.81 to 125 mU/mL are measured with the ScienCell™ Xanthine Oxidase Assay kit. The increase in OD550nm is monitored as a function of time (Figure 1) and the resulting standard curve of ∆OD550nm/min vs xanthine oxidase activity are plotted (Figure 2). -

Xanthine Oxidoreductase in Cancer
Cancer Medicine Open Access REVIEW Xanthine oxidoreductase in cancer: more than a differentiation marker Maria Giulia Battelli, Letizia Polito, Massimo Bortolotti & Andrea Bolognesi Department of Experimental, Diagnostic and Specialty Medicine – DIMES, Alma Mater Studiorum – University of Bologna, General Pathology Unit, Via S. Giacomo 14, 40126 Bologna, Italy Keywords Abstract Differentiation, oncogenesis, reactive oxygen and nitrogen species, uric acid, xanthine Human xanthine oxidoreductase (XOR) catalyzes the last two steps of purine oxidoreductase catabolism and is present in two interconvertible forms, which may utilize O2 or NAD+ as electron acceptors. In addition to uric acid, XOR products may Correspondence comprise reactive oxygen and nitrogen species that have many biologic effects, Letizia Polito, Department of Experimental, including inflammation, endothelial dysfunction, and cytotoxicity, as well as Diagnostic and Specialty Medicine – DIMES, mutagenesis and induction of proliferation. XOR is strictly modulated at the Alma Mater Studiorum – University of Bologna, General Pathology Unit, Via S. transcriptional and post-translational levels, and its expression and activity are Giacomo 14, 40126 Bologna, Italy. highly variable in cancer. Xanthine oxidoreductase (XOR) expression has been Tel: +39 051 2094700; Fax: +39 051 2094746; negatively associated with a high malignity grade and a worse prognosis in E-mail: [email protected] neoplasms of the breast, liver, gastrointestinal tract, and kidney, which normally express a high level of XOR protein. However, the level of XOR expression Funding Information may be associated with a worse outcome in cancer of low XOR-expressing cells, This work was supported by the Pallotti in relation to the inflammatory response elicited through the tissue damage Legacies for Cancer Research. -

Monitoring the Redox Status in Multiple Sclerosis
Preprints (www.preprints.org) | NOT PEER-REVIEWED | Posted: 31 July 2020 doi:10.20944/preprints202007.0737.v1 Review Monitoring the Redox Status in Multiple Sclerosis Masaru Tanaka 1,2 and László Vécsei 1,2,* 1 MTA-SZTE, Neuroscience Research Group, Semmelweis u. 6, Szeged, H-6725 Hungary; [email protected] 2 Department of Neurology, Interdisciplinary Excellence Centre, Faculty of Medicine, University of Szeged, Semmelweis u. 6, H-6725 Szeged, Hungary * Correspondence: [email protected]; Tel.: +36-62-545-351 Received: date; Accepted: date; Published: date Abstract: Worldwide, over 2.2 million people are suffered from multiple sclerosis (MS), a multifactorial demyelinating disease of the central nervous system, characterized by multifocal inflammatory or demyelinating attacks associated with neuroinflammation and neurodegeneration. The blood, cerebrospinal fluid, and postmortem brain samples of MS patients evidenced the presence of reduction-oxidation (redox) homeostasis disturbance such as the alternations of oxidative and antioxidative enzyme activities and the presence of degradation products. This review article discussed the components of redox homeostasis including reactive chemical species, oxidative enzymes, antioxidative enzymes, and degradation products. The reactive chemical species covered frequently discussed reactive oxygen/nitrogen species, rarely featured reactive chemicals such as sulfur, carbonyls, halogens, selenium, and nucleophilic species that potentially act as reductive as well as pro-oxidative stressors. The antioxidative enzyme systems covered the nuclear factor erythroid-2-related factor 2 (NRF2)-Kelch-like ECH-associated protein 1 (KEAP1) signaling pathway, a possible biomarker sensitive to the initial phase of oxidative stress. Altered components of the redox homeostasis in MS were discussed, some of which turned to be MS subtype- or treatment-specific and thus potentially become diagnostic, prognostic, predictive, and/or therapeutic biomarkers. -

Nucleotide Metabolism II
Nucleotide Metabolism II • Biosynthesis of deoxynucleotides • Salvage Pathway • Catabolism: Purines • Catabolism: Pyrimidines • Feedback inhibition in purine nucleotide biosynthesis CPS II • Cytosolic CPS II uses glutamine as the nitrogen donor to carbamoyl phosphate Regulation of pyrimidine synthesis •CPSII is allosterically regulated: PRPP and IMP are activators Several pyrimidines are inhibitors • Aspartate transcarbamoylase (ATCase) Important regulatory point in prokaryotes Catalyzes the first committed pathway step Allosteric regulators: CTP (-), CTP + UTP (-), ATP (+) • Regulation of pyrimidine nucleotide synthesis in E. coli Biosynthesis of deoxynucleotides • Uses diphosphates (ribo) • Ribonucleotide reducatase • 2 sub-units • R1- reduces, active and two allosteric sites (activity and specificity site) • R2- tyrosine radical carries electrons • removes 2' OH to H Ribonucleotide reductase reaction • removes 2' OH to H • Thioredoxin and NADPH used to regenerate sulfhydryl groups Thymidylate synthesis • UDP ------> dUMP • dUMP --------> dTMP • required THF • methylates uracil Regulation THF • Mammals cannot conjugate rings or synthesize PABA. • So must get in diet. • Sulfonamides effective in bacteria due to competitive inhibition of the incorporation of PABA Cancer Drugs • fluorouracil-- suicide inhibitor of Thy synthase • aminopterin • Methotrexate -- inhibits DHF reductase Salvage of Purines and Pyrimidines • During cellular metabolism or digestion, nucleic acids are degraded to heterocyclic bases • These bases can be salvaged -

Diseases Catalogue
Diseases catalogue AA Disorders of amino acid metabolism OMIM Group of disorders affecting genes that codify proteins involved in the catabolism of amino acids or in the functional maintenance of the different coenzymes. AA Alkaptonuria: homogentisate dioxygenase deficiency 203500 AA Phenylketonuria: phenylalanine hydroxylase (PAH) 261600 AA Defects of tetrahydrobiopterine (BH 4) metabolism: AA 6-Piruvoyl-tetrahydropterin synthase deficiency (PTS) 261640 AA Dihydropteridine reductase deficiency (DHPR) 261630 AA Pterin-carbinolamine dehydratase 126090 AA GTP cyclohydrolase I deficiency (GCH1) (autosomal recessive) 233910 AA GTP cyclohydrolase I deficiency (GCH1) (autosomal dominant): Segawa syndrome 600225 AA Sepiapterin reductase deficiency (SPR) 182125 AA Defects of sulfur amino acid metabolism: AA N(5,10)-methylene-tetrahydrofolate reductase deficiency (MTHFR) 236250 AA Homocystinuria due to cystathionine beta-synthase deficiency (CBS) 236200 AA Methionine adenosyltransferase deficiency 250850 AA Methionine synthase deficiency (MTR, cblG) 250940 AA Methionine synthase reductase deficiency; (MTRR, CblE) 236270 AA Sulfite oxidase deficiency 272300 AA Molybdenum cofactor deficiency: combined deficiency of sulfite oxidase and xanthine oxidase 252150 AA S-adenosylhomocysteine hydrolase deficiency 180960 AA Cystathioninuria 219500 AA Hyperhomocysteinemia 603174 AA Defects of gamma-glutathione cycle: glutathione synthetase deficiency (5-oxo-prolinuria) 266130 AA Defects of histidine metabolism: Histidinemia 235800 AA Defects of lysine and -

The Sole Or Key Enzymes of Purine and Pyrimidine Metabolisms Might Direct the Organisms to the Cell Proliferation
EDITORIAL The sole or key enzymes of purine and pyrimidine metabolisms might direct the organisms to the cell proliferation Kristine Edgar Danielyan Danielyan KE. The sole or key enzymes of purine and pyrimidine pathways of purine and pyrimidine metabolism basically are the same. We metabolisms might direct the organisms to the cell proliferation. J Biomol presented over years results, evidencing, inhibition of the cells purine and Biochem 2017;1(1):1-2. pyrimidine catabolism via influence on the key regulative enzymes and ABSTRACT activation of their synthesis might consequently trigger cell proliferation. Cancer development and regenerative cells proliferation are two apposite and Key Words: Purine; Pyrimidine; Metabolism; Cancer; Regeneration; Cells self-excluded processes in the organism. However, the classical biochemical proliferation wo processes – cells regenerative division, including neurogenesis or fast as the regenerative mechanism after experimental stroke. The other face of Tgeneration of epithelial, dermal and other types of the tissues cells and the cells proliferation is the cancer development. proliferation of the cancer cells are two different processes. However, both in the basis have the same classical mechanism of purine and pyrimidine Cancer and regulative enzymes of purine, pyrimidine metabolism biosynthesis as well as their catabolism. Recent data are evidencing, 5-aminoimidazole-4-carboxamide-1-β-riboside In accordance to the rules of biochemical pathways, every chain of the (AICAr) inhibits pyrimidine catabolism and induces multiple myeloma cell death, apoptosis via deprivation of phosphoribosyl pyrophosphate in metabolic interchanges and formations has the regulative components- enzymes, conducting the entire process or pathway. contrast to the impact on the purine metabolism (12). -

O O2 Enzymes Available from Sigma Enzymes Available from Sigma
COO 2.7.1.15 Ribokinase OXIDOREDUCTASES CONH2 COO 2.7.1.16 Ribulokinase 1.1.1.1 Alcohol dehydrogenase BLOOD GROUP + O O + O O 1.1.1.3 Homoserine dehydrogenase HYALURONIC ACID DERMATAN ALGINATES O-ANTIGENS STARCH GLYCOGEN CH COO N COO 2.7.1.17 Xylulokinase P GLYCOPROTEINS SUBSTANCES 2 OH N + COO 1.1.1.8 Glycerol-3-phosphate dehydrogenase Ribose -O - P - O - P - O- Adenosine(P) Ribose - O - P - O - P - O -Adenosine NICOTINATE 2.7.1.19 Phosphoribulokinase GANGLIOSIDES PEPTIDO- CH OH CH OH N 1 + COO 1.1.1.9 D-Xylulose reductase 2 2 NH .2.1 2.7.1.24 Dephospho-CoA kinase O CHITIN CHONDROITIN PECTIN INULIN CELLULOSE O O NH O O O O Ribose- P 2.4 N N RP 1.1.1.10 l-Xylulose reductase MUCINS GLYCAN 6.3.5.1 2.7.7.18 2.7.1.25 Adenylylsulfate kinase CH2OH HO Indoleacetate Indoxyl + 1.1.1.14 l-Iditol dehydrogenase L O O O Desamino-NAD Nicotinate- Quinolinate- A 2.7.1.28 Triokinase O O 1.1.1.132 HO (Auxin) NAD(P) 6.3.1.5 2.4.2.19 1.1.1.19 Glucuronate reductase CHOH - 2.4.1.68 CH3 OH OH OH nucleotide 2.7.1.30 Glycerol kinase Y - COO nucleotide 2.7.1.31 Glycerate kinase 1.1.1.21 Aldehyde reductase AcNH CHOH COO 6.3.2.7-10 2.4.1.69 O 1.2.3.7 2.4.2.19 R OPPT OH OH + 1.1.1.22 UDPglucose dehydrogenase 2.4.99.7 HO O OPPU HO 2.7.1.32 Choline kinase S CH2OH 6.3.2.13 OH OPPU CH HO CH2CH(NH3)COO HO CH CH NH HO CH2CH2NHCOCH3 CH O CH CH NHCOCH COO 1.1.1.23 Histidinol dehydrogenase OPC 2.4.1.17 3 2.4.1.29 CH CHO 2 2 2 3 2 2 3 O 2.7.1.33 Pantothenate kinase CH3CH NHAC OH OH OH LACTOSE 2 COO 1.1.1.25 Shikimate dehydrogenase A HO HO OPPG CH OH 2.7.1.34 Pantetheine kinase UDP- TDP-Rhamnose 2 NH NH NH NH N M 2.7.1.36 Mevalonate kinase 1.1.1.27 Lactate dehydrogenase HO COO- GDP- 2.4.1.21 O NH NH 4.1.1.28 2.3.1.5 2.1.1.4 1.1.1.29 Glycerate dehydrogenase C UDP-N-Ac-Muramate Iduronate OH 2.4.1.1 2.4.1.11 HO 5-Hydroxy- 5-Hydroxytryptamine N-Acetyl-serotonin N-Acetyl-5-O-methyl-serotonin Quinolinate 2.7.1.39 Homoserine kinase Mannuronate CH3 etc. -

Ameen Alsaras Batool Bdour
36 Batool bdour Ameen Alsaras ... Diala Abu Hassan In the previous lecture we started talking about nucleotides, and in this one we’ll talk about how they’re synthesized from scratch ! I. Purine synthesis: *Purines are bigger, they’re made of two connected rings, which indicates that they require a longer pathway to be synthesized than pyrimidines* ▪ The contributing compounds: i. 3 kinds of Amino acids (aspartic acid, glycine and glutamine) ii. CO2 (from the surrounding decarboxylation reactions) iii. N10–formyltetrahydrofolate *you’re not required to know where each atom came from* The purine ring is constructed primarily in the liver. There are actually two ways to synthesize a nucleotide, either de novo where it’s made from scratch and each atom is added to the ring separately *de novo is the major source of DNA synthesis purines* or Salvaged, where big pieces (like the nitrogenous ring) are priorly synthesized and we just put them together. ▪ The steps of synthesis (de novo): We start with the preformed molecule Ribose-5-phosphate, and the build up is accomplished by the serial addition of donated nitrogens and carbons. Ribose 5-phosphate is synthesized by the pentose phosphate pathway * You’re only required to know the names of the main ones like PRPP, IMP. you’re not required to know the order of the steps * Just don’t forget that the pentagon ring was formed first. 1 | P a g e Step 1: activation of synthesis by the addition of pyrophosphate to R-5-P ▪ Activation of of R-5-P occurs by the addition of a pyrophosphate from an ATP molecule by the following equation ATP= PPi + AMP. -
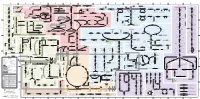
Fullsubwaymap221.Pdf
A B C D E F G H I J K L M Aldose reductase Sorbitol dehydrogenase Cholesterol synthesis Glycogen and galactose metabolism (branch point only) (debranching) glucose sorbitol fructose Aromatic amino acid metabolism Purine synthesis Pyrimidine synthesis glycogen (n+1) phenylacetate triiodothyronine (T3) dopaquinone melanin + + thyroxine (T4) (multiple steps) (many cycles) P 4' NADPH NADP NAD NADH acetyl-CoA x2 i Debranching enzyme 3' - α-1,4 bonds Aromatic amino acid 5-phosphoribosyl pyrophosphate (PRPP) HCO B 2' P 3 (branching) Glycogen 6 i (multiple steps) O H O (multiple steps) Tyrosinase O H O 1' 2 2 2 2 B 1 2 3 4 5 6 7 8 9 10 Pentose phosphate pathway Phenylalanine Tyrosine decarboxylase 6 Dopamine B Thiolase phosphorylase -5 -4 -3 -2 -1 0 1 2 3 4 Transaminase 6 glutamine 2 ATP (many cycles) Glycogen Glucose-6-phosphatase Dehydrogenase hydroxylase hydroxylase (DOPA decarboxylase) β-hydroxylase Methyltransferase 10 UDP 4:4 Transferase ATP glutathione (reduced) H O phenyllactate phenylpyruvate phenylalanine tyrosine dihydroxy- dopamine Glutamine PRPP CoA synthase Hexokinase or (in endoplasmic reticulum) 2 2 norepinephrine epinephrine glutamine Carbamoyl 9 1' phenylanine amidotransferase (GPAT) glutamate glycogen (n) Glutathione reductase Glutathione peroxidase + phosphate glycogen (n) -5 -4 -3 -2 -1 0 1 2 3 4 2' 3' 4' glucokinase 8 NADP NADPH glutamate α-ketoglutarate CO2 O H O Glycosyl (4:6) UDP-glucose ADP (L-DOPA) 2 2 synthetase II glutathione (oxidized) 2 H O α-ketoglutarate PPi glutamate acetoacetyl-CoA 7 1 Glucose -1,6-Glucosidase -

The Role of Metals in Enzyme Activity*
ANNALS OF CLINICAL AND LABORATORY SCIENCE, Vol. 7, No. 2 Copyright © 1977, Institute for Clinical Science The Role of Metals in Enzyme Activity* JAMES F. RIORDAN, Ph.D. Biophysics Research Laboratory, Department of Biological Chemistry, Harvard Medical School, and Division of Medical Biology, Veter Bent Brigham Hospital, Boston, MA 02115 ABSTRACT Metal ions play important roles in the biological function of many en zymes. The various modes of metal-protein interaction include metal-, ligand-, and enzyme-bridge complexes. Metals can serve as electron donors or acceptors, Lewis acids or structural regulators. Those that participate directly in the catalytic mechanism usually exhibit anomalous physicochemical characteristics reflecting their entatic state. Carboxypep- tidase A, liver alcohol dehydrogenase, aspartate transcarbamoylase and al kaline phosphatase exemplify the different roles of metals in metalloen- zymes while the nucleotide polymerases point to the essential role of zinc in maintaining normal growth and development. Introduction At present, reliable measurements of small concentrations of metals present in Certain metals have long been recog tissues, cells, subcellular particles, body nized to have important biological func fluids and biomacromolecules can be tions primarily as a consequence of nu performed by colorimetry, fluorimetry, tritional investigations.14,15,22 Thus, the polarography, emission spectrometry absence of a specific, essential metal with spark, flame or plasma excitation from the diet of an organism