Breeds of Beef and Multi-Purpose Cattle
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Breed Relationships and Definition in British Cattle
Heredity (2004) 93, 597–602 & 2004 Nature Publishing Group All rights reserved 0018-067X/04 $30.00 www.nature.com/hdy Breed relationships and definition in British cattle: a genetic analysis P Wiener, D Burton and JL Williams Roslin Institute (Edinburgh), Roslin, Midlothian EH25 9PS, UK The genetic diversity of eight British cattle breeds was not associated with geographical distribution. Analyses also quantified in this study. In all, 30 microsatellites from the FAO defined the cohesiveness or definition of the various breeds, panel of markers were used to characterise the DNA with Highland, Guernsey and Jersey as the best defined and samples from nearly 400 individuals. A variety of methods most distinctive of the breeds. were applied to analyse the data in order to look at diversity Heredity (2004) 93, 597–602. doi:10.1038/sj.hdy.6800566 within and between breeds. The relationships between Publishedonline25August2004 breeds were not highly resolved and breed clusters were Keywords: British cattle; breeds; diversity; microsatellites Introduction 1997; MacHugh et al, 1994, 1998; Kantanen et al, 2000; Arranz et al, 2001; Bjrnstad and Red 2001; Beja-Pereira The concept of cattle breeds, rather than local types, is et al, 2003). said to have originated in Britain under the influence of The goal of this study was to use microsatellite Robert Bakewell in the 18th century (Porter, 1991). It was markers to characterise diversity levels within, and during that period that intensive culling and inbreeding relationships between, a number of British cattle breeds, became widespread in order to achieve specific breeding most of which have not been characterised previously. -

Beef Showmanship Parts of a Steer
Beef Showmanship Parts of a Steer Wholesale Cuts of a Market Steer Common Cattle Breeds Angus (English) Maine Anjou Charolaise Short Horn Hereford (English) Simmental Showmanship Terms/Questions Bull: an intact adult male Steer: a male castrated prior to development of secondary sexual characteristics Stag: a male castrated after development of secondary sexual characteristics Cow: a female that has given birth Heifer: a young female that has not yet given birth Calf: a young bovine animal Polled: a beef animal that naturally lacks horns 1. What is the feed conversion ratio for cattle? a. 7 lbs. feed/1 lb. gain 2. About what % of water will a calf drink of its body weight in cold weather? a. 8% …and in hot weather? a. 19% 2. What is the average daily weight gain of a market steer? a. 2.0 – 4 lbs./day 3. What is the approximate percent crude protein that growing cattle should be fed? a. 12 – 16% 4. What is the most common concentrate in beef rations? a. Corn 5. What are three examples of feed ingredients used as a protein source in a ration? a. Cottonseed meal, soybean meal, distillers grain brewers grain, corn gluten meal 6. Name two forage products used in a beef cattle ration: a. Alfalfa, hay, ground alfalfa, leaf meal, ground grass 7. What is the normal temperature of a cow? a. 101.0°F 8. The gestation period for a cow is…? a. 285 days (9 months, 7 days) 9. How many stomachs does a steer have? Name them. a. 4: Rumen, Omasum, Abomasum, and Reticulum 10. -

CATAIR Appendix
CBP and Trade Automated Interface Requirements Appendix: PGA April 24, 2020 Pub # 0875-0419 Contents Table of Changes ............................................................................................................................................4 PG01 – Agency Program Codes .................................................................................................................... 18 PG01 – Government Agency Processing Codes ............................................................................................. 22 PG01 – Electronic Image Submitted Codes.................................................................................................... 26 PG01 – Globally Unique Product Identification Code Qualifiers .................................................................... 26 PG01 – Correction Indicators* ...................................................................................................................... 26 PG02 – Product Code Qualifiers.................................................................................................................... 28 PG04 – Units of Measure .............................................................................................................................. 30 PG05 – Scie nt if ic Spec ies Code .................................................................................................................... 31 PG05 – FWS Wildlife Description Codes ..................................................................................................... -

Report Name: Livestock and Products Semi-Annual
Required Report: Required - Public Distribution Date: March 06,2020 Report Number: AR2020-0007 Report Name: Livestock and Products Semi-annual Country: Argentina Post: Buenos Aires Report Category: Livestock and Products Prepared By: Kenneth Joseph Approved By: Melinda Meador Report Highlights: Argentine beef exports in 2020 are projected down at 640,000 tons carcass weight equivalent as lower prices and animal and human health issues generate negative trade dynamics. Lower exports will be reflected in marginal growth expansion of the domestic market in 2020. FAS/USDA has changed the conversion rates for Argentine beef exports. THIS REPORT CONTAINS ASSESSMENTS OF COMMODITY AND TRADE ISSUES MADE BY USDA STAFF AND NOT NECESSARILY STATEMENTS OF OFFICIAL U.S. GOVERNMENT POLICY Conversion Rates: Due to continuing efforts to improve data reliability, the “New Post” trade forecasts reflect new conversion rates. Historical data revisions (from 2005 onward) will be published on April 9th in the Production, Supply and Demand (PSD) database (http://www.fas.usda.gov/psdonline). Beef and Veal Conversion Factors Code Description Conversion Rate* 020110 Bovine carcasses and half carcasses, fresh or chilled 1.0 020120 Bovine cuts bone in, fresh or chilled 1.0 020130 Bovine cuts boneless, fresh or chilled 1.36 020210 Bovine carcasses and half carcasses, frozen 1.0 020220 Bovine cuts bone in, frozen 1.0 020230 Bovine cuts boneless, frozen 1.36 021020 Bovine meat salted, dried or smoked 1.74 160250 Bovine meat, offal nes, not livers, prepared/preserve 1.79 -

Saturday 1St February 2020 All Entered Animals Lotted and Penned As Per Catalogue and Late Entries on a First Come First Served Basis on the Day
LIVESTOCK ENTRIES FOR Saturday 1st February 2020 All entered animals lotted and penned as per catalogue and late entries on a first come first served basis on the day. 9.30am PRIME HOGGS & CAST EWES 10am BREEDING & STORE PIGS 10.30am BREEDING & STORE CATTLE 10.30am BREEDING & STORE SHEEP Inc. In-lamb sheep & with lambs at foot & store hoggs CAST SHEEP/GOATS & PRIME HOGGS 9.30am Start As Forward on the Day BREEDING & STORE PIG SALE 10am start Entries are forward on the day with fortnightly entries of between 100-200 This week includes: SS Perry 20 Large White x Prime pigs LJ Pounder, Bedale 7 `Berkshire stores 5 Large White stores T & G McGarrell, Earby 1 Large White Boar 1 Large White Gilt All prospective pig purchasers and vendors, please ensure you are registered to do so. http://www.eaml2.org.uk/ BREEDING & STORE CATTLE 10.30 am in the Main Ring Full Registered Name Required of all Named Sired Cattle BREEDING BULLS as forward LOT NO. NAME QTY DESCRIPTION TB FA 1000 W Tomlinson, Wycoller 1 Saler stock bull, non-reg 10yo 4 N 999 TW Pickard & Son, GtHarwood 1 Pedigree reg 4yo Limousin stock bull 4 N Procters Farm bred COWS/HEIFERS IN CALF & WITH CALVES LOT NO. NAME QTY DESCRIPTION AGE TB FA 901-10 JP Stansfield Ltd, Todmorden 5 Blonde cows with Blonde calves at foot 4 N 1+1 Limousin heifer 2½yo PD’d 4m back in calf 4 911-12 AJ Maude to Lim “Lodge Hamlet” with Lim bull calf 6m by same bull. -

Crossbreeding of Cattle in Africa
Journal of Agriculture and Environmental Sciences June 2018, Vol. 7, No. 1, pp. 16-31 ISSN: 2334-2404 (Print), 2334-2412 (Online) Copyright © The Author(s). All Rights Reserved. Published by American Research Institute for Policy Development DOI: 10.15640/jaes.v7n1a3 URL: https://doi.org/10.15640/jaes.v7n1a3 Crossbreeding of Cattle in Africa R Trevor Wilson1 Abstract Africa is endowed with a very wide range of mostly Bos indicus indigenous cattle breeds. A general statement with regard to their performance for meat or milk is that they are of inferior genetic value. Attempts to improve their performance have rarely relied on within-breed improvement but have concentrated on crossing to supposedly superior exotic Bos taurus types. Exotic types have not always – indeed have rarely -- been chosen on objective criteria and the imported breeds generally indicate the colonial past of individual African countries rather than on use of “the right animal in the right place”. Most attempts at increasing output have been undertaken under research station conditions. Results on station have been very variable but the limited success achieved has rarely been carried over in to the general African cattle population. This paper documents a number of attempts to alter the genetic make-up of African cattle in several countries and discusses the reasons for the failure of most of these. Keywords: Bos indicus, Bos taurus, livestock experiments, milk production, meat production 1. Introduction African countries differ greatly in climatic, ecological and agricultural conditions and in socioeconomic factors. In many countries, nonetheless, cattle are the most important livestock species. -

Review on Potential of Reproductive Technology to Improved Ruminant Production in Ethiopia
Journal of Biology, Agriculture and Healthcare www.iiste.org ISSN 2224-3208 (Paper) ISSN 2225-093X (Online) Vol.10, No.8, 2020 Review on Potential of Reproductive Technology to Improved Ruminant Production in Ethiopia Jalel Fikadu Yadeta Assosa University, College of Agriculture and Natural Resources, Department of animal science, Assosa, Ethiopia DOI: 10.7176/JBAH/10-8-02 Publication date: April 30 th 2020 INTRODUCTION Reproductive technology encompasses all current and anticipated uses of technology in human and animal reproduction, including assisted reproductive technology, contraception and others (Mapletoft and Hasler, 2005). Research into physiology and embryology has provided a basis for the development of technologies that increase productivity of farm animals through enhanced control of reproductive function. Animal Biotechnology represents an expanding collection of rapidly developing disciplines in science and information technologies. The livestock provides many opportunities to utilize these disciplines and evolving competencies. Individually, these are powerful tools capable of providing significant improvements in productivity. Combinations of these technologies coupled with information systems and data analysis will provide even more significant changes in the next decade. Various techniques have been developed and refined to obtain a large number of offspring from genetically superior animals or obtain offspring from infertile (or sub fertile) animals (Naqvi et al ., 2001; Blackburn, 2004). Based on the progress in scientific knowledge of endocrinology, reproductive physiology, cell biology and embryology during the last fifty years new bio techniques have been developed for and introduced into animal breeding and husbandry (Wrathall et al., 2004). Among them are estrus synchronization/induction, artificial insemination, Multiple Ovulation Induction and Embryo Transfer (MOET), in vitro embryo production (IVP) and cloning by Nuclear Transfer (NT) all are components of the tool box for present and future applications (Betteridge, 2003). -

Type and Breed Characteristics and Uses
E-190 3/09 Texas Adapted Genetic Strategies for Beef Cattle V: Type and Breed Characteristics and Uses A 1700s painting of the foundation cow of one of the first cattle breeds. Courtesy of Michigan State University Animal Science Department. Stephen P. Hammack* he subject of breeds intrigues most beef cattle sphere. The Bos taurus in the United States originated in producers. However, breeds are only part of a ge- the British Isles and western continental Europe. The Bos netic strategy that should include: indicus arose in south central Asia. T• Matching applicable performance or functional There are some intermediates containing both Bos levels to environmental, management, and taurus and Bos indicus. Some intermediates created in the marketing conditions United States, particularly in Texas, are commonly re- • Choosing a breeding system, either continuous ferred to as American breeds, which will be discussed later. (in which replacement females are produced Although it has no strict definition, a breed can be within the herd) or terminal (in which replace- described as animals of common origin with certain dis- ments are introduced externally) tinguishing characteristics that are passed from parent • Selecting genetic types, breeds within types, and individuals within breeds that are compat- *Professor and Extension Beef Cattle Specialist–Emeritus, The Texas A&M System ible with the performance level needed and breeding system chosen. Genetic classifications and breeds Cattle can be divided into two basic classifications, Bos taurus (non-humped) and Bos indicus (humped, also called Zebu). Cattle are not native to the western hemi- to offspring. Breed characteristics result from To characterize milking potential accurately, both natural selection and from that imposed it should be evaluated relative to body size. -

Multiple Choice Choose the Answer That Best Completes Each Statement Or Question
Name Date Hour 5 The Beef Cattle Industry Multiple Choice Choose the answer that best completes each statement or question. _______ 1. Early settlers primarily used cattle as ____ . A. work animals B. a source of meat C. a source of milk D. symbols of wealth _______ 2. The modern cattle industry is concentrated in the ____ . A. South and Midwest B. South and Southwest C. North and Northwest D. North and Midwest _______ 3. Cattle drives were necessary in the past because of the lack of ____ . A. retail stores B. refrigeration C. slaughterhouses D. year-round grazing _______ 4. Which beef cattle breed is known for its solid black color and excellent meat quality? A. Angus B. Hereford C. Shorthorn D. Chianina _______ 5. Which beef cattle breed is from northern England and was often called a Durham after the county in which it originated? A. Angus B. Hereford C. Shorthorn D. Chianina Introduction to Agriscience | Unit 5 Test CIMC 1 _______ 6. Which beef cattle breed originated in England and was originally much larger, weighing more than 3,000 pounds, than it is today? A. Angus B. Hereford C. Shorthorn D. Chianina _______ 7. Which beef cattle breed is red with a white face and may also have white on the neck, underline, legs, and tail switch? A. Angus B. Hereford C. Shorthorn D. Chianina _______ 8. Which beef cattle breed is one of the oldest breeds in the world and originated in Italy? A. Angus B. Hereford C. Shorthorn D. Chianina _______ 9. Which beef cattle breed originated in central France and was developed as a dual-purpose breed and is typically white or off-white in color? A. -

First Report on the State of the World's Animal Genetic Resources"
"First Report on the State of the World’s Animal Genetic Resources" (SoWAnGR) Country Report of the United Kingdom to the FAO Prepared by the National Consultative Committee appointed by the Department for Environment, Food and Rural Affairs (Defra). Contents: Executive Summary List of NCC Members 1 Assessing the state of agricultural biodiversity in the farm animal sector in the UK 1.1. Overview of UK agriculture. 1.2. Assessing the state of conservation of farm animal biological diversity. 1.3. Assessing the state of utilisation of farm animal genetic resources. 1.4. Identifying the major features and critical areas of AnGR conservation and utilisation. 1.5. Assessment of Animal Genetic Resources in the UK’s Overseas Territories 2. Analysing the changing demands on national livestock production & their implications for future national policies, strategies & programmes related to AnGR. 2.1. Reviewing past policies, strategies, programmes and management practices (as related to AnGR). 2.2. Analysing future demands and trends. 2.3. Discussion of alternative strategies in the conservation, use and development of AnGR. 2.4. Outlining future national policy, strategy and management plans for the conservation, use and development of AnGR. 3. Reviewing the state of national capacities & assessing future capacity building requirements. 3.1. Assessment of national capacities 4. Identifying national priorities for the conservation and utilisation of AnGR. 4.1. National cross-cutting priorities 4.2. National priorities among animal species, breeds, -
Proc1-Beginning Chapters.Pmd
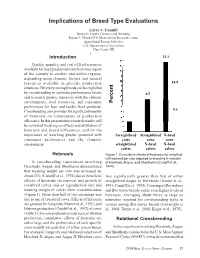
Implications of Breed Type Evaluations Larry V. Cundiff Research Leader, Genetics and Breeding Roman L. Hruska U.S. Meat Animal Research Center Agricultural Research Service U.S. Department of Agriculture Clay Center, NE ()(,) Introduction 23.3 Quality, quantity, and cost of feed resources available for beef production vary from one region of the country to another and within regions, depending upon climatic factors and natural 23.3 resources available in specific production 14.8 situations. Diversity among breeds can be exploited by crossbreeding to optimize performance levels 8.5 and to match genetic resources with the climatic environment, feed resources, and consumer Percent preferences for lean and tender beef products. 8.5 Crossbreeding also provides for significant benefits 14.8 of heterosis on components of production efficiency. In this presentation, research results will be reviewed focusing on effects and utilization of 8.5 8.5 heterosis and breed differences, and on the importance of matching genetic potential with Straightbred Straightbred X-bred consumer preferences and the climatic cows cows cows environment. straightbred X-bred X-bred calves calves calves Heterosis Figure 1. Cumulative effects of heterosis for weight of calf weaned per cow exposed to breeding in crosses A crossbreeding experiment involving of Hereford, Angus, and Shorthorns (Cundiff et al., Herefords, Angus, and Shorthorns demonstrated 1974). that weaning weight per cow was increased by about 23% (Cundiff et al., 1974) due to beneficial was significantly greater than that of either effects of heterosis on survival and growth of straightbred Angus or Herefords (Nunez et al., crossbred calves and on reproduction rate and 1991; Cundiff et al., 1992). -

Prion Protein Gene (PRNP) Variants and Evidence for Strong Purifying Selection in Functionally Important Regions of Bovine Exon 3
Prion protein gene (PRNP) variants and evidence for strong purifying selection in functionally important regions of bovine exon 3 Christopher M. Seabury†, Rodney L. Honeycutt†‡, Alejandro P. Rooney§, Natalie D. Halbert†, and James N. Derr†¶ †Department of Veterinary Pathobiology, College of Veterinary Medicine, Texas A&M University, College Station, TX 77843-4467; ‡Department of Wildlife and Fisheries Sciences, Texas A&M University, College Station, TX 77843-2258; and §National Center for Agricultural Utilization Research, Agricultural Research Service, U.S. Department of Agriculture, Peoria, IL 61604-3999 Communicated by James E. Womack, Texas A&M University, College Station, TX, September 1, 2004 (received for review December 19, 2003) Amino acid replacements encoded by the prion protein gene indel polymorphism has not been observed within the octapep- (PRNP) have been associated with transmissible and hereditary tide repeat region of ovine PRNP exon 3 (8, 10–20), whereas spongiform encephalopathies in mammalian species. However, an studies of cattle and other bovine species have yielded three indel association between bovine spongiform encephalopathy (BSE) and isoforms possessing five to seven octapeptide repeats (20–31). bovine PRNP exon 3 has not been detected. Moreover, little is Despite the importance of cattle both to agricultural practices currently known regarding the mechanisms of evolution influenc- worldwide and to the global economy, surprisingly little is known ing the bovine PRNP gene. Therefore, in this study we evaluated about PRNP allelic diversity for cattle collectively and͞or how the patterns of nucleotide variation associated with PRNP exon 3 this gene evolves in this lineage. In addition, although several for 36 breeds of domestic cattle and representative samples for 10 nondomesticated species of Bovinae contracted transmissible additional species of Bovinae.