Prion Protein Gene (PRNP) Variants and Evidence for Strong Purifying Selection in Functionally Important Regions of Bovine Exon 3
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Boselaphus Tragocamelus</I>
University of Nebraska - Lincoln DigitalCommons@University of Nebraska - Lincoln USGS Staff -- Published Research US Geological Survey 2008 Boselaphus tragocamelus (Artiodactyla: Bovidae) David M. Leslie Jr. U.S. Geological Survey, [email protected] Follow this and additional works at: https://digitalcommons.unl.edu/usgsstaffpub Leslie, David M. Jr., "Boselaphus tragocamelus (Artiodactyla: Bovidae)" (2008). USGS Staff -- Published Research. 723. https://digitalcommons.unl.edu/usgsstaffpub/723 This Article is brought to you for free and open access by the US Geological Survey at DigitalCommons@University of Nebraska - Lincoln. It has been accepted for inclusion in USGS Staff -- Published Research by an authorized administrator of DigitalCommons@University of Nebraska - Lincoln. MAMMALIAN SPECIES 813:1–16 Boselaphus tragocamelus (Artiodactyla: Bovidae) DAVID M. LESLIE,JR. United States Geological Survey, Oklahoma Cooperative Fish and Wildlife Research Unit and Department of Natural Resource Ecology and Management, Oklahoma State University, Stillwater, OK 74078-3051, USA; [email protected] Abstract: Boselaphus tragocamelus (Pallas, 1766) is a bovid commonly called the nilgai or blue bull and is Asia’s largest antelope. A sexually dimorphic ungulate of large stature and unique coloration, it is the only species in the genus Boselaphus. It is endemic to peninsular India and small parts of Pakistan and Nepal, has been extirpated from Bangladesh, and has been introduced in the United States (Texas), Mexico, South Africa, and Italy. It prefers open grassland and savannas and locally is a significant agricultural pest in India. It is not of special conservation concern and is well represented in zoos and private collections throughout the world. DOI: 10.1644/813.1. -

Vega Etal Procroyalsocb Synchronous Diversification
Canterbury Christ Church University’s repository of research outputs http://create.canterbury.ac.uk Please cite this publication as follows: Frantz, Laurent A. F., Rudzinski, A., Mansyursyah Surya Nugraha, A., Evin, A., Burton, J., Hulme-Beaman, A., Linderholm, A., Barnett, R., Vega, R., Irving-Pease, E., Haile, J., Allen, R., Leus, K., Shephard, J., Hillyer, M., Gillemot, S., van den Hurk, J., Ogle, S., Atofanei, C., Thomas, M., Johansson, F., Haris Mustari, A., Williams, J., Mohamad, K., Siska Damayanti, C., Djuwita Wiryadi, I., Obbles, D., Mona, S., Day, H., Yasin, M., Meker, S., McGuire, J., Evans, B., von Rintelen, T., Hoult, S., Searle, J., Kitchener, A., Macdonald, A., Shaw, D., Hall, R., Galbusera, P. and Larson, G. (2018) Synchronous diversification of Sulawesi’s iconic artiodactyls driven by recent geological events. Proceedings of the Royal Society B: Biological Sciences. Link to official URL (if available): http://dx.doi.org/10.1098/rspb.2017.2566. This version is made available in accordance with publishers’ policies. All material made available by CReaTE is protected by intellectual property law, including copyright law. Any use made of the contents should comply with the relevant law. Contact: [email protected] Synchronous diversification of Sulawesi’s iconic artiodactyls driven by recent geological events Authors Laurent A. F. Frantz1,2,a,*, Anna Rudzinski3,*, Abang Mansyursyah Surya Nugraha4,c,*, , Allowen Evin5,6*, James Burton7,8*, Ardern Hulme-Beaman2,6, Anna Linderholm2,9, Ross Barnett2,10, Rodrigo Vega11 Evan K. Irving-Pease2, James Haile2,10, Richard Allen2, Kristin Leus12,13, Jill Shephard14,15, Mia Hillyer14,16, Sarah Gillemot14, Jeroen van den Hurk14, Sharron Ogle17, Cristina Atofanei11, Mark G. -
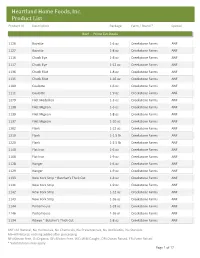
Customer Product List
Heartland Home Foods, Inc. Product List Product Id Description Package Farm / Brand * Special Beef - Prime Cut Steaks 1126 Bavette 1-6 oz Creekstone Farms ANF 1127 Bavette 1-8 oz Creekstone Farms ANF 1116 Chuck Eye 1-8 oz Creekstone Farms ANF 1117 Chuck Eye 1-12 oz Creekstone Farms ANF 1136 Chuck Filet 1-8 oz Creekstone Farms ANF 1135 Chuck Filet 1-16 oz Creekstone Farms ANF 1109 Coulotte 1-6 oz Creekstone Farms ANF 1111 Coulotte 1-9 oz Creekstone Farms ANF 1279 Filet Medallion 1-3 oz Creekstone Farms ANF 1138 Filet Mignon 1-6 oz Creekstone Farms ANF 1139 Filet Mignon 1-8 oz Creekstone Farms ANF 1137 Filet Mignon 1-10 oz Creekstone Farms ANF 1202 Flank 1-12 oz Creekstone Farms ANF 1219 Flank 1-1.5 lb Creekstone Farms ANF 1220 Flank 1-2.5 lb Creekstone Farms ANF 1140 Flat Iron 1-6 oz Creekstone Farms ANF 1168 Flat Iron 1-9 oz Creekstone Farms ANF 1128 Hanger 1-6 oz Creekstone Farms ANF 1129 Hanger 1-9 oz Creekstone Farms ANF 1133 New York Strip ~ Butcher's Thick Cut 1-8 oz Creekstone Farms ANF 1141 New York Strip 1-9 oz Creekstone Farms ANF 1142 New York Strip 1-12 oz Creekstone Farms ANF 1143 New York Strip 1-16 oz Creekstone Farms ANF 1144 Porterhouse 1-18 oz Creekstone Farms ANF 1146 Porterhouse 1-26 oz Creekstone Farms ANF 1134 Ribeye ~ Butcher's Thick Cut 1-8 oz Creekstone Farms ANF ANF=All Natural, No Hormones, No Chemicals, No Preservatives, No Antibiotics, No Steroids AN=All Natural, nothing added after processing NF=Nitrate Free, O=Organic, GF=Gluten Free, WC=Wild Caught, OR=Ocean Raised, FR=Farm Raised * Substitutions may apply -

The Vermont Journal 11-13-19
Rifle PRSRT STD U.S. POSTAGE Season PAID Holiday Happenings POSTAL CUSTOMER RESIDENTIAL CUSTOMER PERMIT #2 Early Holiday Deadlines Opens N. HAVERHILL, NH See Page 3B ECRWSSEDDMECRWSS See Bottom of Page Nov. 16 FREE Your Local Community Newspaper THE NOVEMBERVermont 13, 2019 | WWW.VERMONTJOURNAL.COM JournaVOLUME 19, ISSUEl 46 Area schools welcome Gov. Scott and community to Veterans Day Assembly BY SHARON HUNTLEY to thank someone for their ser- tell his stories and meet gover- The program, celebrating its The Vermont Journal vice. “You should take the time nors. Scott was the 24th gov- seventh year, is due to the vi- to thank a vet or any member ernor that Walton visited. “It sion and hard work of BRHS LUDLOW, Vt. – The seventh of the military, every chance we was a special day for me,” he Booster Club President An- annual Veterans Day Assem- get, every single day,” he said. said. “It’s so important for you, drea Sanford of Ludlow. Ac- bly, Friday, Nov. 8, at Ludlow He then asked all veterans and the younger generation, to do cording to an introduction Elementary School welcomed those serving in the Military to whatever you can to thank our by Color Guard Commander Gov. Phil Scott as part of their stand to be recognized. vets and listen to their stories of American Legion Post 36, moving program to honoring Scott also paid special at- because they truly are heroes Ned Bowen, Sanford went to veterans and active military tention to those of “the great- that set an example for all of the School Board over seven members. -

Richmond 2015-16
RICHMOND StyleWeekly’s Annual Guide to Richmond 2015-16 FIRST PLACE FIRST PLACE W E E E K L L Y Y T ’ S S READERS’ CHOICE 2015 B E D S N T O O M F R I C H THANK John MacLellan Photos & Design & Photos MacLellan John YOU ANNOUNCING OUR 2015-2016 SEASON! BIKINI BABES, SURFER HUNKS AND GIDGET GOES NUTS! THE JOY AND INEVITABILITY OF LIFE, AMIDST GOOD CHICKEN SOUP AND SOME BRISKET PSYCHO BEACH PARTY LAZARUS SYNDROME RICHMOND! by Charles Busch by Bruce Ward; presented as a part of the city-wide Acts of Faith festival JULY 22 – AUGUST 15, 2015 FEBRUARY 24 – MARCH 19, 2016 For voting ABOUT THE PRESSURES OF FAME, PERFECTION AND BEING BARBRA A COMEDY ABOUT EXPLOITATION AND EMPOWERMENT BUYER & CELLAR BODY AWARENESS by Jonathan Tolins by Annie Baker; a co-production with 5th Wall Theatre Project RICHMOND OCTOBER 7–31, 2015 APRIL 20 – MAY 14, 2016 THE KIDS FROM YOUR FAVORITE HOLIDAY STORIES — GROWN-UP, AND SERVED WITH A TWIST! A MUSICAL TRUE STORY, WITH ALL THE GLITTER POSSIBLE TRIANGLE CHRISTMAS ON THE ROCKS THE BOY FROM OZ Conceived by Rob Ruggiero; written by John Cariani, Jeffrey Hatcher, Jacques Lammare, by Martin Sherman and Nick Enright; PLAYERS Matthew Lombardo, Theresa Rebeck, Edwin Sanchez & Jonathan Tolins based on the life and songs of Peter Allen NOVEMBER 18 – DECEMBER 19, 2015 JUNE 8 – JULY 16, 2016 And make sure you check our web site at www.rtriangle.org for our cabaret your favorite nights, special events, and performers checking in from all over the country! theater company! The 2015-16 Season Is Above, some of our Players (left to right): Ian Page, Anna Grey Hogan, Caleb Supported In Part By Funding From Wade, Tarnée Hudson - We 3 Lizas; Danielle Williams, Liz Earnest - 5 Lesbians Eating A Quiche; Audra Honaker - Angels In America; Andrew Etheredge - Pageant; MEDIA SPONSORS: Matt Shofner, Kylie Clark - Angels In America; Steve Boschen - Pageant; Boomie Pederson - Angels In America; (seated) Jeffrey Cole, Matt Polson - Design for 1300 Altamont Avenue Richmond, VA 23230 Living; Drew Colletti, Ed Hughes - YANK!; Jennie Meharg - Design for Living. -

Ungulate Tag Marketing Update Aza Midyear Conference 2015 Columbia, Sc
UNGULATE TAG MARKETING UPDATE AZA MIDYEAR CONFERENCE 2015 COLUMBIA, SC Brent Huffman - Toronto Zoo Michelle Hatwood - Audubon Species Survival Center RoxAnna Breitigan - Cheyenne Mountain Zoo Species Marketing Original Goals Began in 2011 Goal: Focus institutional interest Need to stop declining trend in captive populations Target: Animal decision makers Easy accessibility 2015 Picked 12 priority species to specifically market for sustainability Postcards mailed to 212 people at 156 institutions Postcards Printed on recycled paper Program Leaders asked to provide feedback Interest Out of the 12 Species… . 8 Program Leaders were contacted by new interested parties in 2014 Sitatunga- posters at AZA meeting Bontebok- Word of mouth, facility contacted TAG Urial- Received Ungulate postcard Steenbok- Program Leader initiated contact Bactrian Wapiti- Received Ungulate postcard Babirusa- Program Leader initiated contact Warty Pigs- WPPH TAG website Arabian Oryx- Word of mouth Results Out of the 12 Species… . 4 Species each gained new facilities Bontebok - 1 Steenbok - 1 Warty Pig - 2 (but lost 1) Babirusa - 4 Moving Forward Out of the 12 Species… . Most SSP’s still have animals available . Most SSP’s are still looking for new institutions . Babirusa- no animals available . Anoa- needs help to work with private sector to get more animals . 170 spaces needed to bring these programs up to population goals Moving Forward Ideas for new promotion? . Continue postcards? Posters? Promotional items? Advertisements? Facebook? Budget? To be announced -

Nutribalance-5000 Nutritional Scale
NutriBalance-5000 Nutritional Scale Carb. Guide Contains over 7000 additional food codes for carbohydrates! oz Max: 11lb d: 0.1oz MR M+ WT 9 Prot 7 8 Cal Sal 0 Tare 6 Fat Carb Col 4 5 Fibr 3 g/oz CLR 2 WT MC 1 How To Use This Manual: This manual provides a cross-reference of carbohydrate codes for the NutriBalance nutritional scale, based on the USDA National Nutrient Database Release 18. When using this manual, only the Carb function of the Nutribalance should be used. All other nutritional buttons such as Fiber, Prot, etc will not display accurate information. 1. To find the Carb Code for a food item, simply use the Acrobat Search function (Ctrl+F or Ctrl+Shift+F). Enter the name of the food item in the Search Field and hit Enter. Give the search time to complete. 2. Once you find your food item in the manual, select your code from the “Code to use” column, or the Code (Fiber Method) column. 3. Place the food item onto the weighing platform and enter the code using the keypad. Now press the Carb button. NOTE: The NutriBalance requires 3-digit input for the code to be accepted. Therefore, if the “Code to use” is 3, you should enter 003, etc. Code to use Code Carbo- Fiber_ Refuse_ Modified ( Fiber hydrt TD Pct Carbs (- Method) fiber) MILK SUBSTITUTES,FLUID,W/ 41 41 6.16 0 0 6.16 LAURIC ACID OIL MILK,WHL,3.25% MILKFAT 85 85 4.52 0 0 4.52 MILK,PRODUCER,FLUID,3.7% 819 819 4.65 0 0 4.65 MILKFAT MILK,RED 819 819 4.68 0 0 4.68 FAT,FLUID,2%MILKFAT,W/ADDED VIT A MILK,RED FAT,FLUID,2% 696 696 4.97 0 0 4.97 MILKFAT,W/ NONFAT MILK SOL&VIT A MILK,RED -

NUTRITIONAL ANALYSIS of ANOA (Bubalus Depressicornis and Bubalus Quarlesi) FOOD PLANTS in TANJUNG PEROPA WILDLIFE RESERVE, SOUTHEAST SULAWESI
Media Konservasi Vol. 16, No. 2 Agustus 2011 : 92 ± 94 NUTRITIONAL ANALYSIS OF ANOA (Bubalus depressicornis and Bubalus quarlesi) FOOD PLANTS IN TANJUNG PEROPA WILDLIFE RESERVE, SOUTHEAST SULAWESI (Analisis Kandungan Nutrisi Pakan Anoa Bubalus spp. di Suaka Margasatwa Tanjung Peropa Sulawesi Tenggara) ABDUL HARIS MUSTARI1) 1)Department of Forest Resources Conservation, Faculty of Forestry Bogor Agricultural University ([email protected]) Diterima 24 Jannuary 2011/Disetujui 27 Juli 2011 ABSTRAK Sebanyak 46 jenis tumbuhan dan dua jenis buah dikumpulkan dari habitat asli anoa (insitu) di Suaka Margasatwa Tanjung Peropa Sulawesi Tenggara. Analisis kandungan nutrisi makanan anoa diketahui dengan menggunakan metode Proximate Analyses. Hasil penelitian menunjukkan bahwa persentase kandungan nutrisi makanan anoa di habitat aslinya bervariasi. Kandungan protein bervariasi 5,58 -21,60 (rataan 12,70; SD 4,34), sementara kandungan serat kasar bervariasi dari 14,68 sampai 62,68 (rataan 36,93; SD 12,07). Persentase Ektrak Ether adalah 0,91-11,5 (rataan 2,38; SD 1,75). Persentase kandungan NFE (Nitrogen-Free Extractives) berkisar 0,76 dan 52,31 (rataan 24,64; SD 15,20), dan kandungan energi kasar adalah 2419-3583 kal/gram (rataan 3093; SD 282,82). Kata kunci: Anoa, Bubalus spp., kandungan nutrisi. INTRODUCTION METHODS Very little is known about the dietary ecology of Study site these animals in their natural habitats because of their This study was conducted in Kalobo Forest of secretive nature and their occupation of the most remote Tanjung Peropa wildlife reserve from 2000 to 2003. The tropical rain forests on the island including lowland wildlife reserve situated between 1220 45‘ ± 1220 45‘ forests, rocky-cliff forests and mountainous forests. -

The Saola Or Spindlehorn Bovid Pseudoryx Nghetinhensis in Laos
ORYX VOL 29 NO 2 APRIL 1995 The saola or spindlehorn bovid Pseudoryx nghetinhensis in Laos George B. Schaller and Alan Rabinowitz In 1992 the discovery of a new bovid, Pseudoryx nghetinhensis, in Vietnam led to speculation that the species also occurred in adjacent parts of Laos. This paper describes a survey in January 1994, which confirmed the presence of P. ngethinhensis in Laos, although in low densities and with a patchy distribution. The paper also presents new information that helps clarify the phylogenetic position of the species. The low numbers and restricted range ofP. ngethinhensis mean that it must be regarded as Endangered. While some admirable moves have been made to protect the new bovid and its habitat, more needs to be done and the authors recommend further conservation action. Introduction area). Dung et al. (1994) refer to Pseudoryx as the Vu Quang ox, but, given the total range of In May 1992 Do Tuoc and John MacKinnon the animal and its evolutionary affinities (see found three sets of horns of a previously un- below), we prefer to call it by the descriptive described species of bovid in the Vu Quang local name 'saola'. Nature Reserve of west-central Vietnam The village of Nakadok, where saola horns (Stone, 1992). The discovery at the end of the were found, lies at the end of the Nakai-Nam twentieth century of a large new mammal in a Theun National Biodiversity Conservation region that had been visited repeatedly by sci- Area (NNTNBCA), which at 3500 sq km is the entific and other expeditions (Delacour and largest of 17 protected areas established by Jabouille, 1931; Legendre, 1936) aroused in- Laos in October 1993. -

The Main Beef Breeds
MODULE 1A PART D: STUDENT FACTSHEET - THE MAIN BEEF BREEDS Breed Characteristics Dam Mature Sire Mature Origin Purpose Weight Weight • Colour black or red • Developed in Scotland from cattle native to Aberdeen Angus Aberdeenshire and Angus 600-900kg 900-1100kg Scotland Terminal sire • Naturally polled • Popular beef breed in tHe United States • A fertile cross between domesticated cattle (Bos Taurus) and American Bison (Bison bison). THe intention of tHe cross is to combine tHe lower fat and cholesterol, cold Beefalo resistance and easy calving qualities of the variable variable UDA Cig Eidion Bison with the docility and higher growth rates of domestic cattle. It is only a Beefalo if the cross is 5/8 ths (37.5%) Bison, if the proportion is higher then its called a Hybrid Bison. • Colour varies from white to black. • Large long body witH double muscle in Hind quarter. British Belgian Blues 800kg 1300kg Terminal sire • HigH saleable meat yield Belgium • Increased dystocia an issue witH tHe breed 1 Breed Characteristics Dam Mature Sire Mature Origin Purpose Weight Weight • White-Tan colour • Ease of calving • HigH growtH rates British Blonde 600 kg 700-1100kg France Terminal sire • Unbroken wHeat coloured • Extended gestation period • Colour creamy wHite througH to wHeat • First continental breed of cattle to be introduced to Great Britain • Initial importation of bulls by dairy Terminal sire and sire for ¾ Charolais 600-900kg 900-1100kg France producers seeking a sire to improve their continental suckler cows. calves’ conformation. • HigH daily liveweigHt gain and improved conformation. • Hardy suckler cow breed Galloway • Suitable for low input systems 400 – 600kg 800 - 950 kg Scotland Native suckler cow breed • Long lived cows • White face and red coat • Easy temperament Hereford 700 – 800kg 1200 – 1500kg Hereford Terminal sire • Main terminal sire breed prior to importation of continental breeds. -

Mixed-Species Exhibits with Pigs (Suidae)
Mixed-species exhibits with Pigs (Suidae) Written by KRISZTIÁN SVÁBIK Team Leader, Toni’s Zoo, Rothenburg, Luzern, Switzerland Email: [email protected] 9th May 2021 Cover photo © Krisztián Svábik Mixed-species exhibits with Pigs (Suidae) 1 CONTENTS INTRODUCTION ........................................................................................................... 3 Use of space and enclosure furnishings ................................................................... 3 Feeding ..................................................................................................................... 3 Breeding ................................................................................................................... 4 Choice of species and individuals ............................................................................ 4 List of mixed-species exhibits involving Suids ........................................................ 5 LIST OF SPECIES COMBINATIONS – SUIDAE .......................................................... 6 Sulawesi Babirusa, Babyrousa celebensis ...............................................................7 Common Warthog, Phacochoerus africanus ......................................................... 8 Giant Forest Hog, Hylochoerus meinertzhageni ..................................................10 Bushpig, Potamochoerus larvatus ........................................................................ 11 Red River Hog, Potamochoerus porcus ............................................................... -

MICHIGAN BEEF PRODUCTION COOPERATIVE EXTENSION SERVICE • MICHIGAN STATE UNIVERSITY Selecting a Breed of Beef Cattle Harlan D
Extension Bulletin E-1755 February 1984 (NEW) 80 cents MICHIGAN BEEF PRODUCTION COOPERATIVE EXTENSION SERVICE • MICHIGAN STATE UNIVERSITY Selecting a Breed of Beef Cattle Harlan D. Ritchie Department of Animal Science Criteria For Choosing A Breed considered good milkers. Angus females are known Selecting a breed or combination of breeds to use for their fertility and ease of calving. The breed is in your beef herd should be based on the following nearly pure for the polled trait and Angus bulls can be criteria: (1) marketability in your area; (2) cost and expected to sire calf crops that are 100% hornless. availability of good seedstock; (3) climate; (4) quantity The dark skin pigment provides some resistance and quality of feedstuffs on your farm; (5) how the against cancer eye and sun-burned udders. breeds used in a crossing program complement one Angus calves fatten quickly and grade Choice at a another; and (6) personal preference. As an example of relatively light weight (1,050 lb.). They possess more climatic adaptability, British breeds are well adapted marbling in the meat than any other breed of cattle, to cold climates, but do not fare as well in sub which means their quality grade (Prime, Choice, tropical regions. Conversely, Brahman blood is need Good, etc.) is often higher than that of other cattle. ed for optimum performance in certain Gulf Coastal For this reason, some packers pay a premium for areas, but is not required in the northern states. Angus or Angus-cross steers. However, feedlot operators sometimes pay less for Angus feeder calves British Breeds because they have a tendency to mature too quickly and become fat at too light a weight.