Drosophila Genes Potentially Involved in Responses to Microbial Infection
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Prokaryotes (Domains Bacteria & Archaea)
2/4/15 Prokaryotes (Domains Bacteria & Archaea) KEY POINTS 1. Decomposers: recycle organic and inorganic molecules in environment; makes them available to other organisms. 2. Essential components of symbioses. 3. Encompasses the origins of metabolism and metabolic diversity. 4. Origin of photosynthesis and formation of atmospheric Oxygen Ceno- Meso- zoic zoic ANTIQUITY Humans Paleozoic Colonization of land Animals Origin of solar system and Earth • >3.5 BILLION years old. • Alone for 2 1 4 billion years Proterozoic Archaean Prokaryotes Billions of 2 years ago3 Multicellular eukaryotes Single-celled eukaryotes Atmospheric oxygen General characteristics 1. Small: compare to 10-100µm for 0.5-5µm eukaryotic cell; single-celled; may form colonies. 2. Lack membrane- enclosed organelles. 3. Cell wall present, but different from plant cell wall. 1 2/4/15 General characteristics 4. Occur everywhere, most numerous organisms. – More individuals in a handful of soil then there are people that have ever lived. – By far more individuals in our gut than eukaryotic cells that are actually us. General characteristics 5. Metabolic diversity established nutritional modes of eukaryotes. General characteristics 6. Important decomposers and recyclers 2 2/4/15 General characteristics 6. Important decomposers and recyclers • Form the basis of global nutrient cycles. General characteristics 7. Symbionts!!!!!!! • Parasites • Pathogenic organisms. • About 1/2 of all human diseases are caused by Bacteria General characteristics 7. Symbionts!!!!!!! • Parasites • Pathogenic organisms. • Extremely important in agriculture as well. Pierce’s disease is caused by Xylella fastidiosa, a Gamma Proteobacteria. It causes over $56 million in damage annually in California. That’s with $34 million spent to control it! = $90 million in California alone. -

A Generic Large-Scale Cause for Platelet Dysfunction and Depletion in Infection
Published online: 2020-04-12 THIEME 302 A Champion of Host Defense: A Generic Large-Scale Cause for Platelet Dysfunction and Depletion in Infection Martin J. Page, BSc (Hons)1 Etheresia Pretorius, PhD1 1 Department of Physiological Sciences, Stellenbosch University, Address for correspondence Etheresia Pretorius, PhD, Department of Stellenbosch, South Africa Physiological Sciences, Faculty of Science, Stellenbosch University, Private Bag X1 Matieland, Stellenbosch, 7602, South Africa Semin Thromb Hemost 2020;46:302–319. (e-mail: [email protected]). Abstract Thrombocytopenia is commonly associated with sepsis and infections, which in turn are characterized by a profound immune reaction to the invading pathogen. Platelets are one of the cellular entities that exert considerable immune, antibacterial, and antiviral actions, and are therefore active participants in the host response. Platelets are sensitive to surrounding inflammatory stimuli and contribute to the immune response by multiple mechanisms, including endowing the endothelium with a proinflammatory phenotype, enhancing and amplifying leukocyte recruitment and inflammation, promoting the effector functions of immune cells, and ensuring an optimal adaptive immune response. During infection, pathogens and their products influence the platelet response and can even be toxic. However, platelets are able to sense and engage bacteria and viruses to assist in their removal and destruction. Platelets greatly contribute to host defense by multiple mechanisms, including forming immune complexes and aggregates, shedding their granular content, and internalizing pathogens and subsequently being marked for removal. These processes, and the nature of platelet function in general, cause the platelet to be irreversibly consumed in Keywords the execution of its duty. An exaggerated systemic inflammatory response to infection ► platelets can drive platelet dysfunction, where platelets are inappropriately activated and face ► virus immunological destruction. -

The Wnt Signaling Pathway Is Involved in the Regulation of Phagocytosis Of
OPEN The Wnt signaling pathway is involved in SUBJECT AREAS: the regulation of phagocytosis of virus in PHAGOCYTES CELL BIOLOGY Drosophila CYTOSKELETON Fei Zhu1,2 & Xiaobo Zhang1 CELLULAR MICROBIOLOGY 1Key Laboratory of Conservation Biology for Endangered Wildlife of Ministry of Education, Key Laboratory of Animal Virology of 2 Received Ministry of Agriculture and College of Life Sciences, Zhejiang University, Hangzhou 310058, China, College of Animal Science 1 February 2013 and Technology, Zhejiang Agriculture and Forestry University, Hangzhou 311300, China. Accepted 3 June 2013 Phagocytosis is crucial for triggering host defenses against invading pathogens in animals. However, the receptors on phagocyte surface required for phagocytosis of virus have not been extensively explored. This Published study demonstrated that white spot syndrome virus (WSSV), a major pathogen of shrimp, could be engulfed 25 June 2013 but not digested by Drosophila S2 cells, indicating that the virus was not recognized and taken up by a pathway that was silent and would not activate the phagosome maturation and digestion pathway. The results showed that the activation of receptors on S2 cell surface by lipopolysaccharide or peptidoglycan resulted in the phagocytosis of S2 cells against WSSV virions. Gene expression profiles revealed that the Correspondence and dally-mediated Wnt signaling pathway was involved in S2 phagocytosis. Further data showed that the Wnt requests for materials signaling pathway played an essential role in phagocytosis. Therefore, our study contributed novel insight should be addressed to into the molecular mechanism of phagocytosis in animals. X.B.Z. (zxb0812@zju. edu.cn) hagocytosis, a highly conserved process, is crucial for the immune responses of animals and it allows for rapid engulfment of pathogens and apoptotic cells by specialized phagocytes1–5. -

Generated by SRI International Pathway Tools Version 25.0, Authors S
An online version of this diagram is available at BioCyc.org. Biosynthetic pathways are positioned in the left of the cytoplasm, degradative pathways on the right, and reactions not assigned to any pathway are in the far right of the cytoplasm. Transporters and membrane proteins are shown on the membrane. Periplasmic (where appropriate) and extracellular reactions and proteins may also be shown. Pathways are colored according to their cellular function. Gcf_000238675-HmpCyc: Bacillus smithii 7_3_47FAA Cellular Overview Connections between pathways are omitted for legibility. -

Dynamic Regulation of Innate Immune Responses in Drosophila by Senju-Mediated Glycosylation
Dynamic regulation of innate immune responses in Drosophila by Senju-mediated glycosylation Miki Yamamoto-Hinoa, Masatoshi Muraokab, Shu Kondoc, Ryu Uedac, Hideyuki Okanod, and Satoshi Gotoa,d,1 aDepartment of Life Science, Rikkyo University, Tokyo 171-8501, Japan; bStem Cell Project Group, Tokyo Metropolitan Institute of Medical Science, Tokyo 156-8506, Japan; cInvertebrate Genetics Laboratory, Genetic Strains Research Center, National Institute of Genetics, Mishima 411-8540, Japan; and dDepartment of Physiology, Keio University School of Medicine, Tokyo 160-8582, Japan Edited by Norbert Perrimon, Howard Hughes Medical Institute, Harvard Medical School, Boston, MA, and approved March 30, 2015 (received for review December 22, 2014) The innate immune system is the first line of defense encountered by Most cell surface and secreted proteins are glycosylated in the invading pathogens. Delayed and/or inadequate innate immune endoplasmic reticulum (ER) and Golgi apparatus. Glycosylation responses can result in failure to combat pathogens, whereas ex- requires glycosyltransferases and nucleotide sugar substrates cessive and/or inappropriate responses cause runaway inflamma- that are synthesized in the cytosol and nucleus. The nucleotide tion. Therefore, immune responses are tightly regulated from sugars must be transported into the ER and Golgi lumens by initiation to resolution and are repressed during the steady state. It is nucleotide sugar transporters (NSTs) (6). On arrival, they are used well known that glycans presented on pathogens play important by glycosyltransferases. The Drosophila genome encodes at least roles in pathogen recognition and the interactions between host 10 NSTs, which transport different subsets of nucleotide sugars. molecules and microbes; however, the function of glycans of host Studies of mutated NST genes show that protein glycosylation organisms in innate immune responses is less well known. -

Killing of Gram-Negative Bacteria by Lactoferrin and Lysozyme
Killing of gram-negative bacteria by lactoferrin and lysozyme. R T Ellison 3rd, T J Giehl J Clin Invest. 1991;88(4):1080-1091. https://doi.org/10.1172/JCI115407. Research Article Although lactoferrin has antimicrobial activity, its mechanism of action is not full defined. Recently we have shown that the protein alters the Gram-negative outer membrane. As this membrane protects Gram-negative cells from lysozyme, we have studied whether lactoferrin's membrane effect could enhance the antibacterial activity of lysozyme. We have found that while each protein alone is bacteriostatic, together they can be bactericidal for strains of V. cholerae, S. typhimurium, and E. coli. The bactericidal effect is dose dependent, blocked by iron saturation of lactoferrin, and inhibited by high calcium levels, although lactoferrin does not chelate calcium. Using differing media, the effect of lactoferrin and lysozyme can be partially or completely inhibited; the degree of inhibition correlating with media osmolarity. Transmission electron microscopy shows that E. coli cells exposed to lactoferrin and lysozyme at 40 mOsm become enlarged and hypodense, suggesting killing through osmotic damage. Dialysis chamber studies indicate that bacterial killing requires direct contact with lactoferrin, and work with purified LPS suggests that this relates to direct LPS-binding by the protein. As lactoferrin and lysozyme are present together in high levels in mucosal secretions and neutrophil granules, it is probable that their interaction contributes to host defense. Find the latest version: https://jci.me/115407/pdf Killing of Gram-negative Bacteria by Lactofernn and Lysozyme Richard T. Ellison III*" and Theodore J. Giehl *Medical and tResearch Services, Department of Veterans Affairs Medical Center, and Division ofInfectious Diseases, Department ofMedicine, University ofColorado School ofMedicine, Denver, Colorado 80220 Abstract (4). -

The Histone Deacetylase HDA15 Interacts with MAC3A and MAC3B to Regulate Intron
bioRxiv preprint doi: https://doi.org/10.1101/2020.11.17.386672; this version posted November 17, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. 1 The histone deacetylase HDA15 interacts with MAC3A and MAC3B to regulate intron 2 retention of ABA-responsive genes 3 4 Yi-Tsung Tu1#, Chia-Yang Chen1#, Yi-Sui Huang1, Ming-Ren Yen2, Jo-Wei Allison Hsieh2,3, 5 Pao-Yang Chen2,3*and Keqiang Wu1* 6 7 1Institute of Plant Biology, National Taiwan University, Taipei 10617, Taiwan 8 2 Institute of Plant and Microbial Biology, Academia Sinica, Taipei 11529, Taiwan 9 3 Genome and Systems Biology Degree Program, Academia Sinica and National Taiwan 10 University, Taipei, Taiwan 11 12 # These authors contributed equally to this work. 13 * Corresponding authors: Keqiang Wu ([email protected], +886-2-3366-4546) and Pao-Yang 14 Chen ([email protected], +886-2-2787-1140) 15 16 Short title: HDA15 and MAC3A/MAC3B coregulate intron retention 17 One sentence summary: HDA15 and MAC3A/MAC3B coregulate intron retention and reduce 18 the histone acetylation level of the genomic regions near ABA-responsive retained introns. 19 20 The author responsible for distribution of materials integral to the findings presented in this 21 article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) 22 is: Keqiang Wu ([email protected]) 23 24 1 bioRxiv preprint doi: https://doi.org/10.1101/2020.11.17.386672; this version posted November 17, 2020. -

Supplementary Table S1. Table 1. List of Bacterial Strains Used in This Study Suppl
Supplementary Material Supplementary Tables: Supplementary Table S1. Table 1. List of bacterial strains used in this study Supplementary Table S2. List of plasmids used in this study Supplementary Table 3. List of primers used for mutagenesis of P. intermedia Supplementary Table 4. List of primers used for qRT-PCR analysis in P. intermedia Supplementary Table 5. List of the most highly upregulated genes in P. intermedia OxyR mutant Supplementary Table 6. List of the most highly downregulated genes in P. intermedia OxyR mutant Supplementary Table 7. List of the most highly upregulated genes in P. intermedia grown in iron-deplete conditions Supplementary Table 8. List of the most highly downregulated genes in P. intermedia grown in iron-deplete conditions Supplementary Figures: Supplementary Figure 1. Comparison of the genomic loci encoding OxyR in Prevotella species. Supplementary Figure 2. Distribution of SOD and glutathione peroxidase genes within the genus Prevotella. Supplementary Table S1. Bacterial strains Strain Description Source or reference P. intermedia V3147 Wild type OMA14 isolated from the (1) periodontal pocket of a Japanese patient with periodontitis V3203 OMA14 PIOMA14_I_0073(oxyR)::ermF This study E. coli XL-1 Blue Host strain for cloning Stratagene S17-1 RP-4-2-Tc::Mu aph::Tn7 recA, Smr (2) 1 Supplementary Table S2. Plasmids Plasmid Relevant property Source or reference pUC118 Takara pBSSK pNDR-Dual Clonetech pTCB Apr Tcr, E. coli-Bacteroides shuttle vector (3) plasmid pKD954 Contains the Porpyromonas gulae catalase (4) -

Natural Products That Target the Arginase in Leishmania Parasites Hold Therapeutic Promise
microorganisms Review Natural Products That Target the Arginase in Leishmania Parasites Hold Therapeutic Promise Nicola S. Carter, Brendan D. Stamper , Fawzy Elbarbry , Vince Nguyen, Samuel Lopez, Yumena Kawasaki , Reyhaneh Poormohamadian and Sigrid C. Roberts * School of Pharmacy, Pacific University, Hillsboro, OR 97123, USA; cartern@pacificu.edu (N.S.C.); stamperb@pacificu.edu (B.D.S.); fawzy.elbarbry@pacificu.edu (F.E.); nguy6477@pacificu.edu (V.N.); lope3056@pacificu.edu (S.L.); kawa4755@pacificu.edu (Y.K.); poor1405@pacificu.edu (R.P.) * Correspondence: sroberts@pacificu.edu; Tel.: +1-503-352-7289 Abstract: Parasites of the genus Leishmania cause a variety of devastating and often fatal diseases in humans worldwide. Because a vaccine is not available and the currently small number of existing drugs are less than ideal due to lack of specificity and emerging drug resistance, the need for new therapeutic strategies is urgent. Natural products and their derivatives are being used and explored as therapeutics and interest in developing such products as antileishmanials is high. The enzyme arginase, the first enzyme of the polyamine biosynthetic pathway in Leishmania, has emerged as a potential therapeutic target. The flavonols quercetin and fisetin, green tea flavanols such as catechin (C), epicatechin (EC), epicatechin gallate (ECG), and epigallocatechin-3-gallate (EGCG), and cinnamic acid derivates such as caffeic acid inhibit the leishmanial enzyme and modulate the host’s immune response toward parasite defense while showing little toxicity to the host. Quercetin, EGCG, gallic acid, caffeic acid, and rosmarinic acid have proven to be effective against Leishmania Citation: Carter, N.S.; Stamper, B.D.; in rodent infectivity studies. -

Elimination of Plasmatocytes by Targeted Apoptosis Reveals Their Role in Multiple Aspects of the Drosophila Immune Response
Elimination of plasmatocytes by targeted apoptosis reveals their role in multiple aspects of the Drosophila immune response Bernard Charroux and Julien Royet1 Institut de Biologie du De´veloppement de Marseille Luminy, Unite´Mixte de Recherche 6216, Centre National de la Recherche Scientifique, Universite´dela Méditerrannée Aix-Marseille II, 13288 Marseille Cedex 9, France Communicated by Jules A. Hoffmann, Centre National de la Recherche Scientifique, Strasbourg, France, April 17, 2009 (received for review January 14, 2009) Drosophila hemocytes have strong phagocytic capacities and pro- generating plasmatocyte-depleted individuals and by analyzing duce antimicrobial peptides (AMPs). However, the precise role of their development and immune response. blood cells during immune responses and developmental processes has only been studied using indirect means. To overcome this Results limitation, we generated plasmatocyte-depleted flies by specifi- To obtain flies devoid of plasmatocytes, we decided to trigger cally overexpressing the proapoptotic protein Hid into plasmato- cell death in blood cells by overexpressing the proapoptotic cytes. Unexpectedly, these plasmatocyte-depleted animals have a protein Hid using the plasmatocyte (and crystal cells)–specific normal larval and pupal development and do not exhibit any hml(⌬)-Gal4 driver (20, 21). A UAS-EGFP transgene was used obvious defect after birth. Remarkably, plasmatocyte-depleted to precisely characterize the spatiotemporal expression of the adults show a strong susceptibility to infections by various micro- hml(⌬)-Gal4 driver. GFP is not expressed during embryogenesis, organisms, although activation of systemic AMP gene transcription even in late embryos in which Croquemort-positive blood cells via the Toll and immune deficiency (IMD) pathways is wild-type. are detected (Fig. -

B Protein Relish Regulates the JNK-Mediated Immune Response in Drosophila
Downloaded from genesdev.cshlp.org on September 24, 2021 - Published by Cold Spring Harbor Laboratory Press Targeting of TAK1 by the NF-B protein Relish regulates the JNK-mediated immune response in Drosophila Jin Mo Park,1 Helen Brady,3 Maria Grazia Ruocco,1 Huaiyu Sun,2 DeeAnn Williams,2 Susan J. Lee,2 Tomohisa Kato Jr.,1 Normand Richards,3 Kyle Chan,3 Frank Mercurio,3 Michael Karin,1 and Steven A. Wasserman2,4 1Laboratory of Gene Regulation and Signal Transduction, Department of Pharmacology, School of Medicine, and 2Center for Molecular Genetics, Section of Cell and Developmental Biology, Division of Biology, University of California, San Diego, La Jolla, California 92093-0636, USA; 3Celgene Corporation, San Diego, California 92121, USA The molecular circuitry underlying innate immunity is constructed of multiple,evolutionarily conserved signaling modules with distinct regulatory targets. The MAP kinases and the IKK-NF-B molecules play important roles in the initiation of immune effector responses. We have found that the Drosophila NF-B protein Relish plays a crucial role in limiting the duration of JNK activation and output in response to Gram-negative infections. Relish activation is linked to proteasomal degradation of TAK1,the upstream MAP kinase kinase kinase required for JNK activation. Degradation of TAK1 leads to a rapid termination of JNK signaling,resulting in a transient JNK-dependent response that precede s the sustained induction of Relish-dependent innate immune loci. Because the IKK-NF-B module also negatively regulates JNK activation in mammals,thereby controlling inflammation-induced apoptosis,the regulatory cross-talk between the JNK and NF-B pathways appears to be broadly conserved. -
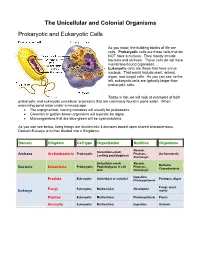
The Unicellular and Colonial Organisms Prokaryotic And
The Unicellular and Colonial Organisms Prokaryotic and Eukaryotic Cells As you know, the building blocks of life are cells. Prokaryotic cells are those cells that do NOT have a nucleus. They mostly include bacteria and archaea. These cells do not have membrane-bound organelles. Eukaryotic cells are those that have a true nucleus. That would include plant, animal, algae, and fungal cells. As you can see, to the left, eukaryotic cells are typically larger than prokaryotic cells. Today in lab, we will look at examples of both prokaryotic and eukaryotic unicellular organisms that are commonly found in pond water. When examining pond water under a microscope… The unpigmented, moving microbes will usually be protozoans. Greenish or golden-brown organisms will typically be algae. Microorganisms that are blue-green will be cyanobacteria. As you can see below, living things are divided into 3 domains based upon shared characteristics. Domain Eukarya is further divided into 4 Kingdoms. Domain Kingdom Cell type Organization Nutrition Organisms Absorb, Unicellular-small; Prokaryotic Photsyn., Archaeacteria Archaea Archaebacteria Lacking peptidoglycan Chemosyn. Unicellular-small; Absorb, Bacteria, Prokaryotic Peptidoglycan in cell Photsyn., Bacteria Eubacteria Cyanobacteria wall Chemosyn. Ingestion, Eukaryotic Unicellular or colonial Protozoa, Algae Protista Photosynthesis Fungi, yeast, Fungi Eukaryotic Multicellular Absorption Eukarya molds Plantae Eukaryotic Multicellular Photosynthesis Plants Animalia Eukaryotic Multicellular Ingestion Animals Prokaryotic Organisms – the archaea, non-photosynthetic bacteria, and cyanobacteria Archaea - Microorganisms that resemble bacteria, but are different from them in certain aspects. Archaea cell walls do not include the macromolecule peptidoglycan, which is always found in the cell walls of bacteria. Archaea usually live in extreme, often very hot or salty environments, such as hot mineral springs or deep-sea hydrothermal vents.