Explosive Radiation in High Andean Hypericum—Rates of Diversification
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

The Vascular Plants of Massachusetts
The Vascular Plants of Massachusetts: The Vascular Plants of Massachusetts: A County Checklist • First Revision Melissa Dow Cullina, Bryan Connolly, Bruce Sorrie and Paul Somers Somers Bruce Sorrie and Paul Connolly, Bryan Cullina, Melissa Dow Revision • First A County Checklist Plants of Massachusetts: Vascular The A County Checklist First Revision Melissa Dow Cullina, Bryan Connolly, Bruce Sorrie and Paul Somers Massachusetts Natural Heritage & Endangered Species Program Massachusetts Division of Fisheries and Wildlife Natural Heritage & Endangered Species Program The Natural Heritage & Endangered Species Program (NHESP), part of the Massachusetts Division of Fisheries and Wildlife, is one of the programs forming the Natural Heritage network. NHESP is responsible for the conservation and protection of hundreds of species that are not hunted, fished, trapped, or commercially harvested in the state. The Program's highest priority is protecting the 176 species of vertebrate and invertebrate animals and 259 species of native plants that are officially listed as Endangered, Threatened or of Special Concern in Massachusetts. Endangered species conservation in Massachusetts depends on you! A major source of funding for the protection of rare and endangered species comes from voluntary donations on state income tax forms. Contributions go to the Natural Heritage & Endangered Species Fund, which provides a portion of the operating budget for the Natural Heritage & Endangered Species Program. NHESP protects rare species through biological inventory, -

The Wetland Condition Index (WCI): Biological Indicators of Wetland Condition for Isolated Depressional Herbaceous Wetlands in Florida
The Wetland Condition Index (WCI): Biological Indicators of Wetland Condition for Isolated Depressional Herbaceous Wetlands in Florida Report Submitted to the Florida Department of Environmental Protection under Contract #WM-683 By Charles R. Lane with Mark T. Brown, Mike Murray-Hudson, and M. Benjamin Vivas H.T. Odum Center for Wetlands University of Florida Gainesville, FL 32611-6350 September 2003 ACKNOWLEDGMENTS This research was supported under a research contract from the Florida Department of Environmental Protection (FDEP); M.T. Brown, principal investigator. We would like to acknowledge the support of FDEP staff, especially Russ Frydenborg, Ashley O’Neal, Ellen McCarron, Liz Miller, Lori Wolfe, Joy Jackson, Johnny Richardson, and the FDEP Central Laboratory in Tallahassee. Additional acknowledgement is due to the research group of the H.T. Odum Center for Wetlands, especially Kelly Reiss, Matt Cohen, Jim Surdick, and Susan Carstenn. We also thank the members of the 1999 and 2000 field crew for their tireless and enthusiastic work, especially Mark Fowlkes, Mark Otto, and Michael Stevens. Steven Doherty was an integral part of the initial stages of this research, and his perseverance and hard work helped to ensure this research to fruition. This project and the preparation of this report were funded in part by a Section 319 Nonpoint Source Management grant from the U.S. Environmental Protection Agency through a contract with the Florida Department of Environmental Protection. ii TABLE OF CONTENTS Page ACKNOWLEDGMENTS ................................................................................................. -

Impacts of Laurel Wilt Disease on Native Persea of the Southeastern United States Timothy M
Clemson University TigerPrints All Dissertations Dissertations 5-2016 Impacts of Laurel Wilt Disease on Native Persea of the Southeastern United States Timothy M. Shearman Clemson University, [email protected] Follow this and additional works at: https://tigerprints.clemson.edu/all_dissertations Recommended Citation Shearman, Timothy M., "Impacts of Laurel Wilt Disease on Native Persea of the Southeastern United States" (2016). All Dissertations. 1656. https://tigerprints.clemson.edu/all_dissertations/1656 This Dissertation is brought to you for free and open access by the Dissertations at TigerPrints. It has been accepted for inclusion in All Dissertations by an authorized administrator of TigerPrints. For more information, please contact [email protected]. IMPACTS OF LAUREL WILT DISEASE ON NATIVE PERSEA OF THE SOUTHEASTERN UNITED STATES A Dissertation Presented to the Graduate School of Clemson University In Partial Fulfillment of the Requirements for the Degree Doctor of Philosophy Forest Resources by Timothy M. Shearman May 2016 Accepted by: Dr. G. Geoff Wang, Committee Chair Dr. Saara J. DeWalt Dr. Donald L. Hagan Dr. Julia L. Kerrigan Dr. William C. Bridges ABSTRACT Laurel Wilt Disease (LWD) has caused severe mortality in native Persea species of the southeastern United States since it was first detected in 2003. This study was designed to document the range-wide population impacts to LWD, as well as the patterns of mortality and regeneration in Persea ecosystems. I used Forest Inventory and Analysis (FIA) data from the U.S. Forest Service to estimate Persea borbonia (red bay) populations from 2003 to 2011 to see if any decline could be observed since the introduction of LWD causal agents. -

National List of Vascular Plant Species That Occur in Wetlands 1996
National List of Vascular Plant Species that Occur in Wetlands: 1996 National Summary Indicator by Region and Subregion Scientific Name/ North North Central South Inter- National Subregion Northeast Southeast Central Plains Plains Plains Southwest mountain Northwest California Alaska Caribbean Hawaii Indicator Range Abies amabilis (Dougl. ex Loud.) Dougl. ex Forbes FACU FACU UPL UPL,FACU Abies balsamea (L.) P. Mill. FAC FACW FAC,FACW Abies concolor (Gord. & Glend.) Lindl. ex Hildebr. NI NI NI NI NI UPL UPL Abies fraseri (Pursh) Poir. FACU FACU FACU Abies grandis (Dougl. ex D. Don) Lindl. FACU-* NI FACU-* Abies lasiocarpa (Hook.) Nutt. NI NI FACU+ FACU- FACU FAC UPL UPL,FAC Abies magnifica A. Murr. NI UPL NI FACU UPL,FACU Abildgaardia ovata (Burm. f.) Kral FACW+ FAC+ FAC+,FACW+ Abutilon theophrasti Medik. UPL FACU- FACU- UPL UPL UPL UPL UPL NI NI UPL,FACU- Acacia choriophylla Benth. FAC* FAC* Acacia farnesiana (L.) Willd. FACU NI NI* NI NI FACU Acacia greggii Gray UPL UPL FACU FACU UPL,FACU Acacia macracantha Humb. & Bonpl. ex Willd. NI FAC FAC Acacia minuta ssp. minuta (M.E. Jones) Beauchamp FACU FACU Acaena exigua Gray OBL OBL Acalypha bisetosa Bertol. ex Spreng. FACW FACW Acalypha virginica L. FACU- FACU- FAC- FACU- FACU- FACU* FACU-,FAC- Acalypha virginica var. rhomboidea (Raf.) Cooperrider FACU- FAC- FACU FACU- FACU- FACU* FACU-,FAC- Acanthocereus tetragonus (L.) Humm. FAC* NI NI FAC* Acanthomintha ilicifolia (Gray) Gray FAC* FAC* Acanthus ebracteatus Vahl OBL OBL Acer circinatum Pursh FAC- FAC NI FAC-,FAC Acer glabrum Torr. FAC FAC FAC FACU FACU* FAC FACU FACU*,FAC Acer grandidentatum Nutt. -
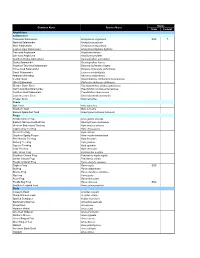
St. Joseph Bay Native Species List
Status Common Name Species Name State Federal Amphibians Salamanders Flatwoods Salamander Ambystoma cingulatum SSC T Marbled Salamander Ambystoma opacum Mole Salamander Ambystoma talpoideum Eastern Tiger Salamander Ambystoma tigrinum tigrinum Two-toed Amphiuma Amphiuma means One-toed Amphiuma Amphiuma pholeter Southern Dusky Salamander Desmognathus auriculatus Dusky Salamander Desmognathus fuscus Southern Two-lined Salamander Eurycea bislineata cirrigera Three-lined Salamander Eurycea longicauda guttolineata Dwarf Salamander Eurycea quadridigitata Alabama Waterdog Necturus alabamensis Central Newt Notophthalmus viridescens louisianensis Slimy Salamander Plethodon glutinosus glutinosus Slender Dwarf Siren Pseudobranchus striatus spheniscus Gulf Coast Mud Salamander Pseudotriton montanus flavissimus Southern Red Salamander Pseudotriton ruber vioscai Eastern Lesser Siren Siren intermedia intermedia Greater Siren Siren lacertina Toads Oak Toad Bufo quercicus Southern Toad Bufo terrestris Eastern Spadefoot Toad Scaphiopus holbrooki holbrooki Frogs Florida Cricket Frog Acris gryllus dorsalis Eastern Narrow-mouthed Frog Gastrophryne carolinensis Western Bird-voiced Treefrog Hyla avivoca avivoca Cope's Gray Treefrog Hyla chrysoscelis Green Treefrog Hyla cinerea Southern Spring Peeper Hyla crucifer bartramiana Pine Woods Treefrog Hyla femoralis Barking Treefrog Hyla gratiosa Squirrel Treefrog Hyla squirella Gray Treefrog Hyla versicolor Little Grass Frog Limnaoedus ocularis Southern Chorus Frog Pseudacris nigrita nigrita Ornate Chorus Frog Pseudacris -

The Quarterly Journal of the Florida Native Plant Society
Volume 28: Number 3 > Summer/Fall 2011 PalmettoThe Quarterly Journal of the Florida Native Plant Society Invasion of the Climbing Ferns ● Wildflowers of Tosohatchee ● Stickywilly ● Landscape Awards Part II Right: Checking the book. Photo by Vince Lamb. Hunting Wildflowers at Tosohatchee WMA Walter Kingsley Taylor The long-anticipated day had arrived. It was Thursday, May 19, and the 31st Annual Conference of the Florida Native Plant Society was a reality. The fruits of labor from all the planning, numerous meetings held, and emails exchanged would come to fruition. Field trip “G” to the Tosohatchee Wildlife Management Area (TOS) near Christmas, Orange County, Florida, was about to roll. Just a few days earlier, the new Taylor Creek Road bridge was finished and opened for traffic. This cut traveling miles for the 20 folks scheduled to come on trip “G”, and the other TOS FNPS field trip led by Katherine Bowman and Pete Dunkelberg. The weather was perfect. Fortunately, the area had recently received a needed rain and I knew the plants would be perked-up and ready to show off their colors. About a week before this gorgeous day, Karin and I motored to the TOS to survey things only to find the ground to be quite dry with many plants drooping. As we drove through the TOS main gate, there were our good friends Ray Jarrett, Sid Taylor, and Rita Grant, and a few other attendees to greet us. Ray had his boots on and was ready to go. He had directed the first vehicles to the appropriate parking area. -

Assessment Report on Hypericum Perforatum L., Herba
European Medicines Agency Evaluation of Medicines for Human Use London, 12 November 2009 Doc. Ref.: EMA/HMPC/101303/2008 COMMITTEE ON HERBAL MEDICINAL PRODUCTS (HMPC) ASSESSMENT REPORT ON HYPERICUM PERFORATUM L., HERBA 7 Westferry Circus, Canary Wharf, London, E14 4HB, UK Tel. (44-20) 74 18 84 00 Fax (44-20) 75 23 70 51 E-mail: [email protected] http://www.emea.europa.eu © European Medicines Agency, 2009. Reproduction is authorised provided the source is acknowledged TABLE OF CONTENTS I. REGULATORY STATUS OVERVIEW...................................................................................4 II. ASSESSMENT REPORT............................................................................................................5 II.1 INTRODUCTION..........................................................................................................................6 II.1.1 Description of the herbal substance(s), herbal preparation(s) or combinations thereof 6 II.1.1.1 Herbal substance:........................................................................................................ 6 II.1.1.2 Herbal preparation(s): ................................................................................................ 7 II.1.1.3 Combinations of herbal substance(s) and/or herbal preparation(s)........................... 9 Not applicable. ................................................................................................................................9 II.1.1.4 Vitamin(s) ................................................................................................................... -

Field Identification Guide to WAP Plants 2008 3Rd Ed
The Field Identification Guide to Plants Used in the Wetland Assessment Procedure (WAP) Contributors: Shirley R. Denton, Ph.D. - Biological Research Associates Diane Willis, MS – GPI Southeast, Inc. April 2008 Third Edition (2015 Printing) The Field Identification Guide was prepared by the Southwest Florida Water Management District. Additional copies can be obtained from the District at: Southwest Florida Water Management District Resource Projects Department Ecological Evaluation Section 2379 Broad Street Brooksville, Florida 34604 The Southwest Florida Water Management District (District) does not discriminate on the basis of disability. This nondiscrimination policy involves every aspect of the District’s functions, including access to and participation in the District’s programs and activities. Anyone requiring reasonable accommodation as provided for in the Americans with Disabilities Act should contact the District’s Human Resources Bureau Chief, 2379 Broad St., Brooksville, FL 34604-6899; telephone (352) 796-7211 or 1-800-423-1476 (FL only), ext. 4703; or email [email protected]. If you are hearing or speech impaired, please contact the agency using the Florida Relay Service, 1(800)955-8771 (TDD) or 1(800)955-8770 (Voice). Introduction In 1996, the Florida Legislature directed the Southwest Florida Water Management District (District) to begin the process of establishing Minimum Flows and Levels (MFLs) throughout the District, beginning in Hillsborough, Pasco, and Pinellas counties. MFLs are defined as the flow in watercourses below which significant harm to water resources and ecology of the area would occur, and the level in surface-water bodies and aquifers in which significant harm to the water resources of the area would occur. -

Sommaire Liste Des Tableaux
SOMMAIRE LISTE DES TABLEAUX ............................................................................................................................ II LISTE DES FIGURES ............................................................................................................................... III LISTE DES PLANCHES PHOTOS ............................................................................................................... III LISTE DES ABREVIATIONS UTILISEES .................................................................................................... IV A. LA BIODIVERSITE TERRESTRE .......................................................................................... 1 A.1 LA BIODIVERSITE VEGETALE............................................................................................ 1 A.1.1 FLORE SPONTANEE ................................................................................................................ 1 A.1.1.1 Flore autochtone .............................................................................................................. 1 A.1.1.1.1 Richesse spécifique ................................................................................................................... 1 A.1.1.1.2 Espèces endémiques ................................................................................................................. 5 A.1.1.1.3 Espèces très rares, rares, assez rares ....................................................................................... 13 A.1.1.1.4 Espèces menacées -

ATLAS of FLORIDA PLANTS - 7/29/19 Lake County Native Species
ATLAS OF FLORIDA PLANTS - 7/29/19 Lake County Native Species Scientific_Name Common_Name Endemic State US 1 Abutilon hulseanum MAUVE N 2 Acalypha gracilens SLENDER THREESEED MERCURY N 3 Acalypha ostryifolia PINELAND THREESEED MERCURY N 4 Acer negundo BOXELDER N 5 Acer rubrum RED MAPLE N 6 Acrolejeunea heterophylla 7 Acrostichum danaeifolium GIANT LEATHER FERN N 8 Aeschynomene americana SHYLEAF N 9 Aeschynomene viscidula STICKY JOINTVETCH N 10 Aesculus pavia RED BUCKEYE N 11 Agalinis fasciculata BEACH FALSE FOXGLOVE N 12 Agalinis filifolia SEMINOLE FALSE FOXGLOVE N 13 Agalinis linifolia FLAXLEAF FALSE FOXGLOVE N 14 Agalinis plukenetii PLUKENET'S FALSE FOXGLOVE N 15 Agarista populifolia FLORIDA HOBBLEBUSH; PIPESTEM N 16 Ageratina jucunda HAMMOCK SNAKEROOT N 17 Aletris lutea YELLOW COLICROOT N 18 Allium canadense var. canadense MEADOW GARLIC N 19 Amaranthus australis SOUTHERN AMARANTH N 20 Amblystegium serpens 21 Ambrosia artemisiifolia COMMON RAGWEED N 22 Amorpha fruticosa BASTARD FALSE INDIGO N 23 Amorpha herbacea var. herbacea CLUSTERSPIKE FALSE INDIGO N 24 Amphicarpum muehlenbergianum BLUE MAIDENCANE N 25 Amsonia ciliata FRINGED BLUESTAR N 26 Andropogon brachystachyus SHORTSPIKE BLUESTEM N 27 Andropogon floridanus FLORIDA BLUESTEM N 28 Andropogon glomeratus var. glaucopsis PURPLE BLUESTEM N 29 Andropogon glomeratus var. hirsutior BUSHY BLUESTEM N 30 Andropogon glomeratus var. pumilus BUSHY BLUESTEM N 31 Andropogon gyrans ELLIOTT'S BLUESTEM N 32 Andropogon longiberbis HAIRY BLUESTEM N 33 Andropogon ternarius SPLITBEARD BLUESTEM N 34 Andropogon -

Toxicity Assessment of Hypericum Olympicum Subsp. Olympicum L. On
J Appl Biomed journal homepage: http://jab.zsf.jcu.cz DOI: 10.32725/jab.2020.002 Journal of Applied Biomedicine Original research article Toxicity assessment of Hypericum olympicum subsp. olympicum L. on human lymphocytes and breast cancer cell lines Necmiye Balikci 1, Mehmet Sarimahmut 1, Ferda Ari 1, Nazlihan Aztopal 1, 2, Mustafa Zafer Özel 3, Engin Ulukaya 1, 4, Serap Celikler 1 * 1 Uludag University, Faculty of Science and Arts, Department of Biology, Bursa, Turkey 2 Istinye University, Faculty of Science and Literature, Department of Molecular Biology and Genetics, Istanbul, Turkey 3 University of York, Department of Chemistry, Heslington, York, United Kingdom 4 Istinye University, Faculty of Medicine, Department of Medical Biochemistry, Istanbul, Turkey Abstract There is a limited number of studies about the constituents ofHypericum olympicum subsp. olympicum and its genotoxic and cytotoxic potency. We examined the possible antigenotoxic/genotoxic properties of methanolic extract of H. olympicum subsp. olympicum (HOE) on human lymphocytes by employing sister chromatid exchange, micronucleus and comet assay and analyzed its chemical composition by GCxGC-TOF/MS. The anti-growth activity against MCF-7 and MDA-MB-231 cell lines was assessed by using the ATP viability assay. Cell death mode was investigated with fluorescence staining and ELISA assays. The major components of the flower and trunk were determined as eicosane, heptacosane, 2-propen-1-ol, hexahydrofarnesyl acetone and α-muurolene. HOE caused significant DNA damage at selected doses (250–750 µg/ml) while chromosomal damage was observed at higher concentrations (500 and 750 µg/ml). HOE demonstrated anti-growth activity in a dose-dependent manner between 3.13–100 µg/ml. -

Response of Two Chrysolina Species to Different Hypericum Hosts
Seventeenth Australasian Weeds Conference Response of two Chrysolina species to different Hypericum hosts Ronny Groenteman', Simon V. Fowler' and Jon J. Sullivan2 ' Landcare Research, Gerald Street, PO Box 40, Lincoln, New Zealand 2Bio-Protection Research Centre, PO Box 84, Lincoln University, Lincoln, New Zealand Corresponding author: [email protected] SummaryChrysolina hyperici and C. quadrigemina introduced in 1963 but, for many years was thought (Coleoptera: Chrysomelidae) were introduced to to have failed to establish (reviewed by Hancox et al. New Zealand for biological control of St John's wort 1986). Chrysolina quadrigemina was rediscovered in (SJW), Hypericum perforatum, following successful the late 1980s (Fraser and Emberson 1987). It is now biological control in Australia. In other parts of the abundant in mixed populations with C. hyperici (R. invaded range of SJW worldwide C. quadrigemina is Groenteman personal observations). generally accepted as the more significant contributor St John's wort beetles are not strictly restricted to to SJW successful biocontrol. Their ability to feed and H. perforatum, and are known to be able to develop on develop on indigenous Hypericum species was not other Hypericum species (some examples are reviewed tested. Chrysolina hyperici established well while C. by Harris 1988). New Zealand hosts 10 naturalised quadrigemina was initially thought to have failed to es- (Healy 1972) and four indigenous (Heenan 2008) tablish in New Zealand, although it is now widespread. Hypericum species. Little is known about the suit- Thus, identifying differences between Chrysolina spe- ability and impacts of either Chrysolina species on cies in host preference and performance would have these Hypericum species.