NIH Public Access Author Manuscript Arthritis Rheum
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

The Ligands for Human Igg and Their Effector Functions
antibodies Review The Ligands for Human IgG and Their Effector Functions Steven W. de Taeye 1,2,*, Theo Rispens 1 and Gestur Vidarsson 2 1 Sanquin Research, Dept Immunopathology and Landsteiner Laboratory, Amsterdam UMC, University of Amsterdam, 1066 CX Amsterdam, The Netherlands; [email protected] 2 Sanquin Research, Dept Experimental Immunohematology and Landsteiner Laboratory, Amsterdam UMC, University of Amsterdam, 1066 CX Amsterdam, The Netherlands; [email protected] * Correspondence: [email protected] Received: 26 March 2019; Accepted: 18 April 2019; Published: 25 April 2019 Abstract: Activation of the humoral immune system is initiated when antibodies recognize an antigen and trigger effector functions through the interaction with Fc engaging molecules. The most abundant immunoglobulin isotype in serum is Immunoglobulin G (IgG), which is involved in many humoral immune responses, strongly interacting with effector molecules. The IgG subclass, allotype, and glycosylation pattern, among other factors, determine the interaction strength of the IgG-Fc domain with these Fc engaging molecules, and thereby the potential strength of their effector potential. The molecules responsible for the effector phase include the classical IgG-Fc receptors (FcγR), the neonatal Fc-receptor (FcRn), the Tripartite motif-containing protein 21 (TRIM21), the first component of the classical complement cascade (C1), and possibly, the Fc-receptor-like receptors (FcRL4/5). Here we provide an overview of the interactions of IgG with effector molecules and discuss how natural variation on the antibody and effector molecule side shapes the biological activities of antibodies. The increasing knowledge on the Fc-mediated effector functions of antibodies drives the development of better therapeutic antibodies for cancer immunotherapy or treatment of autoimmune diseases. -

Annual Report 2015
McGill University Department of Microbiology and Immunology 2015 Annual Report Submitted by J. Madrenas, MD, PhD, FCAHS Canada Research Chair in Human Immunology Professor and Chairman On behalf of Primary members of the Department of Microbiology and Immunology Section I Introduction A. Executive Summary 2015 was an exciting year in the life of the Department of Microbiology and Immunology at McGill. During 2015, we had an External Cyclical Review of the Department, the first review of its kind for (at least) the last 22 years. This process identified key strengths and challenges for our unit, and coalesced different strategic initiatives in consultation with the leadership in the Faculty of Medicine and the Provost office. In 2015, the department welcomed Dr. Corinne Maurice arriving from Harvard University. Corinne started as a tenure track assistant professor on January 1. Her arrival was the fourth recruitment in the context of departmental faculty renewal in the last three years. In addition, we completed the selection process to recruit a Virologist to the Department: the selected candidate was Dr. Jacques Archambault, who will be joining on August 1, 2016 as a Full Professor with tenure. We successfully completed the promotion to Full Professors of Drs. Ciriacco Piccirillo and Donald Sheppard and the 3 yr. reappointment process for Dr. Selena Sagan, and initiated the promotion to Associate Professor with tenure for Dr. Jorg Fritz (a process successfully completed in May 2016). In Research, our members were very successful in attracting high calibre awards. Dr. Mark Wainberg was a 2016 Canadian Medical Hall of Fame inductee. Dr. -

Genetic Variations in the PSMA6 and PSMC6 Proteasome Genes Are Associated with Multiple Sclerosis and Response to Interferon‑Β Therapy in Latvians
EXPERIMENTAL AND THERAPEUTIC MEDICINE 21: 478, 2021 Genetic variations in the PSMA6 and PSMC6 proteasome genes are associated with multiple sclerosis and response to interferon‑β therapy in Latvians NATALIA PARAMONOVA1, JOLANTA KALNINA1, KRISTINE DOKANE1, KRISTINE DISLERE1, ILVA TRAPINA1, TATJANA SJAKSTE1 and NIKOLAJS SJAKSTE1,2 1Genomics and Bioinformatics, Institute of Biology of The University of Latvia; 2Department of Medical Biochemistry of The University of Latvia, LV‑1004 Riga, Latvia Received July 8, 2020; Accepted December 8, 2020 DOI: 10.3892/etm.2021.9909 Abstract. Several polymorphisms in genes related to the Introduction ubiquitin‑proteasome system exhibit an association with pathogenesis and prognosis of various human autoimmune Multiple sclerosis (MS) is a lifelong demyelinating disease of diseases. Our previous study reported the association the central nervous system. The clinical onset of MS tends to between multiple sclerosis (MS) and the PSMA3‑rs2348071 be between the second and fourth decade of life. Similarly to polymorphism in the Latvian population. The current study other autoimmune diseases, women are affected 3‑4 times more aimed to evaluate the PSMA6 and PSMC6 genetic variations, frequently than men (1). About 10% of MS patients experience their interaction between each other and with the rs2348071, a primary progressive MS form characterized by the progres‑ on the susceptibility to MS risk and response to therapy in sion of neurological disability from the onset. In about 90% the Latvian population. PSMA6‑rs2277460, ‑rs1048990 and of MS patients, the disease undergoes the relapse‑remitting PSMC6‑rs2295826, ‑rs2295827 were genotyped in the MS MS course (RRMS); in most of these patients, the condition case/control study and analysed in terms of genotype‑protein acquires secondary progressive course (SPMS) (2). -

Supplementary Table 1: Adhesion Genes Data Set
Supplementary Table 1: Adhesion genes data set PROBE Entrez Gene ID Celera Gene ID Gene_Symbol Gene_Name 160832 1 hCG201364.3 A1BG alpha-1-B glycoprotein 223658 1 hCG201364.3 A1BG alpha-1-B glycoprotein 212988 102 hCG40040.3 ADAM10 ADAM metallopeptidase domain 10 133411 4185 hCG28232.2 ADAM11 ADAM metallopeptidase domain 11 110695 8038 hCG40937.4 ADAM12 ADAM metallopeptidase domain 12 (meltrin alpha) 195222 8038 hCG40937.4 ADAM12 ADAM metallopeptidase domain 12 (meltrin alpha) 165344 8751 hCG20021.3 ADAM15 ADAM metallopeptidase domain 15 (metargidin) 189065 6868 null ADAM17 ADAM metallopeptidase domain 17 (tumor necrosis factor, alpha, converting enzyme) 108119 8728 hCG15398.4 ADAM19 ADAM metallopeptidase domain 19 (meltrin beta) 117763 8748 hCG20675.3 ADAM20 ADAM metallopeptidase domain 20 126448 8747 hCG1785634.2 ADAM21 ADAM metallopeptidase domain 21 208981 8747 hCG1785634.2|hCG2042897 ADAM21 ADAM metallopeptidase domain 21 180903 53616 hCG17212.4 ADAM22 ADAM metallopeptidase domain 22 177272 8745 hCG1811623.1 ADAM23 ADAM metallopeptidase domain 23 102384 10863 hCG1818505.1 ADAM28 ADAM metallopeptidase domain 28 119968 11086 hCG1786734.2 ADAM29 ADAM metallopeptidase domain 29 205542 11085 hCG1997196.1 ADAM30 ADAM metallopeptidase domain 30 148417 80332 hCG39255.4 ADAM33 ADAM metallopeptidase domain 33 140492 8756 hCG1789002.2 ADAM7 ADAM metallopeptidase domain 7 122603 101 hCG1816947.1 ADAM8 ADAM metallopeptidase domain 8 183965 8754 hCG1996391 ADAM9 ADAM metallopeptidase domain 9 (meltrin gamma) 129974 27299 hCG15447.3 ADAMDEC1 ADAM-like, -

Rabbit Anti-Phospho-PTPN6-SL5578R-FITC
SunLong Biotech Co.,LTD Tel: 0086-571- 56623320 Fax:0086-571- 56623318 E-mail:[email protected] www.sunlongbiotech.com Rabbit Anti-phospho-PTPN6 SL5578R-FITC Product Name: Anti-phospho-PTPN6(Ser591)/FITC Chinese Name: FITC标记的磷酸化蛋白酪氨酸磷酸酶1C抗体 PTPN6(phospho S591); SHP1(phospho S591); p-SHP1(Ser591); SHP-1(Phospho- Tyr591); PTPN6(phospho Y591); PTPN6(phospho Y591); SHP1; 70 kda SHP 1L protein; 70 kda SHP1L protein; 70Z-SHP; EC 3.1.3.48; HCP; HCPH; Hematopoietic cell phosphatase; Hematopoietic cell protein tyrosine phosphatase; Hematopoietic cell protein-tyrosine phosphatase; HPTP 1C; HPTP1C; MGC124580; Protein tyrosine Alias: phosphatase 1C; Protein tyrosine phosphatase non receptor type 6; Protein tyrosine phosphatase SHP 1; Protein tyrosine phosphatase SHP1; Protein-tyrosine phosphatase 1C; Protein-tyrosine phosphatase SHP-1; PTN6_HUMAN; PTP 1C; PTP-1C; PTP1C; PTPN 6; PTPN6; SH PTP 1; SH PTP1; SH-PTP1; SHP 1; SHP 1L; SHP1L; SHPTP 1; SHPTP1; Tyrosine protein phosphatase non receptor type 6; Tyrosine-protein phosphatase non-receptor type 6. Organism Species: Rabbit Clonality: Polyclonal React Species: Human,Mouse,Rat,Dog,Pig,Horse, Flow-Cyt=1:50-200IF=1:50-200www.sunlongbiotech.com Applications: not yet tested in other applications. optimal dilutions/concentrations should be determined by the end user. Molecular weight: 65kDa Form: Lyophilized or Liquid Concentration: 1mg/ml KLH conjugated Synthesised phosphopeptide derived from human PTPN6 around the immunogen: phosphorylation site of Ser591 [KG(p-S)LK] Lsotype: IgG Purification: affinity purified by Protein A Storage Buffer: 0.01M TBS(pH7.4) with 1% BSA, 0.03% Proclin300 and 50% Glycerol. Store at -20 °C for one year. Avoid repeated freeze/thaw cycles. -

Disease Phenotypes and Gender Association of FCRL3 Single
Disease Phenotypes and Gender Association of FCRL3 Single-Nucleotide Polymorphism –169T/C in Taiwanese Patients with Systemic Lupus Erythematosus and Rheumatoid Arthritis JI-YIH CHEN, CHIN-MAN WANG, YEONG-JIAN JAN WU, SHIN-NING KUO, CHIUNG-FANG SHIU, SU-WEI CHANG, YEN-TSUN LIN, HUEI-HUANG HO, and JIANMING WU ABSTRACT. Objective. To investigate the association of the functional FCRL3 single-nucleotide polymorphism (SNP) –169T/C with disease phenotypes and susceptibility to systemic lupus erythematosus (SLE) and rheumatoid arthritis (RA) in Taiwanese. Methods. FCRL3 SNP –169T/C was genotyped in 573 patients with SLE, 670 patients with RA, and 758 controls. Genotype distributions and allele frequencies were compared among the 3 groups as aggregates or as stratified by clinical characteristics, autoantibody profile, and sex within patient groups. Results. Overall, FCRL3 SNP –169T/C was not associated with susceptibility to either SLE or RA. However, –169CC genotype was significantly reduced in leukopenia-positive SLE patients as com- pared to the leukopenia-negative SLE patients (CC vs CT+TT, p = 6 × 10–4, OR 0.444, 95% CI 0.279–0.708) and controls (p = 6.1 × 10–3, OR 0.583, 95% CI 0.396–0.857). On the other hand, –169TT genotypes were significantly more numerous in RA patients with non-destructive disease as compared with patients with destructive disease (CC+CT vs TT: p = 0.007, OR 1.672, 95% CI 1.149–2.432). The –169T allele frequency was also significantly increased in non-destructive RA compared with patients with destructive disease (C vs T: p = 0.010, OR 1.423, 95% CI 1.089–1.859). -

Nº Ref Uniprot Proteína Péptidos Identificados Por MS/MS 1 P01024
Document downloaded from http://www.elsevier.es, day 26/09/2021. This copy is for personal use. Any transmission of this document by any media or format is strictly prohibited. Nº Ref Uniprot Proteína Péptidos identificados 1 P01024 CO3_HUMAN Complement C3 OS=Homo sapiens GN=C3 PE=1 SV=2 por 162MS/MS 2 P02751 FINC_HUMAN Fibronectin OS=Homo sapiens GN=FN1 PE=1 SV=4 131 3 P01023 A2MG_HUMAN Alpha-2-macroglobulin OS=Homo sapiens GN=A2M PE=1 SV=3 128 4 P0C0L4 CO4A_HUMAN Complement C4-A OS=Homo sapiens GN=C4A PE=1 SV=1 95 5 P04275 VWF_HUMAN von Willebrand factor OS=Homo sapiens GN=VWF PE=1 SV=4 81 6 P02675 FIBB_HUMAN Fibrinogen beta chain OS=Homo sapiens GN=FGB PE=1 SV=2 78 7 P01031 CO5_HUMAN Complement C5 OS=Homo sapiens GN=C5 PE=1 SV=4 66 8 P02768 ALBU_HUMAN Serum albumin OS=Homo sapiens GN=ALB PE=1 SV=2 66 9 P00450 CERU_HUMAN Ceruloplasmin OS=Homo sapiens GN=CP PE=1 SV=1 64 10 P02671 FIBA_HUMAN Fibrinogen alpha chain OS=Homo sapiens GN=FGA PE=1 SV=2 58 11 P08603 CFAH_HUMAN Complement factor H OS=Homo sapiens GN=CFH PE=1 SV=4 56 12 P02787 TRFE_HUMAN Serotransferrin OS=Homo sapiens GN=TF PE=1 SV=3 54 13 P00747 PLMN_HUMAN Plasminogen OS=Homo sapiens GN=PLG PE=1 SV=2 48 14 P02679 FIBG_HUMAN Fibrinogen gamma chain OS=Homo sapiens GN=FGG PE=1 SV=3 47 15 P01871 IGHM_HUMAN Ig mu chain C region OS=Homo sapiens GN=IGHM PE=1 SV=3 41 16 P04003 C4BPA_HUMAN C4b-binding protein alpha chain OS=Homo sapiens GN=C4BPA PE=1 SV=2 37 17 Q9Y6R7 FCGBP_HUMAN IgGFc-binding protein OS=Homo sapiens GN=FCGBP PE=1 SV=3 30 18 O43866 CD5L_HUMAN CD5 antigen-like OS=Homo -

Replication of Reported Genetic Associations of PADI4, FCRL3
J Hum Genet (2008) 53:163–173 DOI 10.1007/s10038-007-0232-4 ORIGINAL ARTICLE Replication of reported genetic associations of PADI4, FCRL3, SLC22A4 and RUNX1 genes with rheumatoid arthritis: results of an independent Japanese population and evidence from meta-analysis of East Asian studies Yoichiro Takata Æ Hiroshi Inoue Æ Aya Sato Æ Kazue Tsugawa Æ Katsutoshi Miyatake Æ Daisuke Hamada Æ Fumio Shinomiya Æ Shunji Nakano Æ Natsuo Yasui Æ Toshihito Tanahashi Æ Mitsuo Itakura Received: 30 October 2007 / Accepted: 19 November 2007 / Published online: 18 December 2007 Ó The Japan Society of Human Genetics and Springer 2007 Abstract We conducted population-based association magnitudes of effects were apparently much weaker than tests for the four selected SNPs (rs2240340/padi4_94, those reported in the initial positive reports, and there were rs7528684/fcrl3_3, rs3792876/slc2F2 and rs2268277/ substantial levels of inter-study OR heterogeneity, we runx1) previously reported to be associated with rheuma- concluded that additional studies are needed to fully toid arthritis (RA). The study population consisted of 950 understand the present results. unrelated Japanese subjects with RA and 507 controls, none of whom had previously been tested for these vari- Keywords Rheumatoid arthritis Á PADI4 Á FCRL3 Á ants. Only the SNP rs2240340/padi4_94 was modestly SLC22A4 Á RUNX1 Á Single nucleotide polymorphisms Á associated with RA [allele odds ratio (OR) 1.22, 95% Genetic association study Á Meta-analysis confidence interval (CI) 1.04–1.43, P = 0.012]. The most significant association effect was found for genotype con- trast between minor and major allele homozygotes (OR Introduction 1.53, 95% CI 1.10–2.12, P = 0.010). -
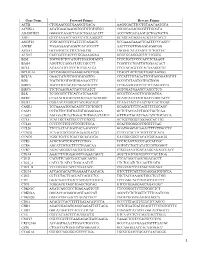
Gene Name Forward Primer Reverse Primer ACTB
Gene Name Forward Primer Reverse Primer ACTB CTGGAACGGTGAAGGTGACA AAGGGACTTCCTGTAACAATGCA ACVRL1 ACATGAAGAAGGTGGTGTGTGTGG CGGGCAGAGGGGTTTGGGTA ADAMDEC1 GGGGCCAGACTACACTGAAACATT ACCCGTCACAAGTACTGATGCTG AHI1 GTCCAAAACTACCCCATCAAGGCT GCAGCACAGGAACGTATCACCT ANGPT2 TGGCAGCGTTGATTTTCAGAGG GCGAAACAAACTCATTTCCCAGCC ANPEP TGAAGAAGCAGGTCACACCCCT AACTCCGTTGGAGCAGGCGG APOA1 GCCGTGCTCTTCCTGACGG TGGGACACATAGTCTCTGCCGC ATXN7 CACCGCCCACTCTGGAAAAGAA GGGTGCAGGGCTTCTTGGTG B2M TGCTGTCTCCATGTTTGATGTATCT TCTCTGCTCCCCACCTCTAAGT BAG4 AGGTTCCAGGATATCCGCCTT TCGGTCCTGATTGTGGAACACT BCL2 ACAACATCGCCCTGTGGATGA CCGTACAGTTCCACAAAGGCAT BCL2L14 GCTCAGGGTCAAAGGACGTTGG TCAGCTACTCGGTTGGCAATGG BCL7A GAACCATGTCGGGCAGGTCG CCCATTTGTAGATTCGTAGGGATGTGT BIN1 TGCTGTCGTGGTGGAGACCTTC GCCGTGTAGTCGTGCTGGG BIRC3 TGCTATCCACATCAGACAGCCC TCTGAATGGTCTTCTCCAGGTTCA BIRC5 TTCTCAAGGACCACCGCATCT AGTGGATGAAGCCAGCCTCG BLK TCGGGGTCTTCACCATCAAAGC GCGCTCCAGGTTGCGGATGA BTRC CCAAATGTGTCATTACCAACATGGGC GCAGCACATAGTGATTTGGCATCC BUB3 CGGAACATGGGTTACGTGCAGC CCAAATACTCAACTGCCACTCGGC CAGE1 TCCAAAATGCACAGTCTTCTGGCT GGAGGCTCTTCAGTTTTTGCAGC CASP1 CCTGTTCCTGTGATGTGGAGGAAA GCTCTACCATCTGGCTGCTCAA CASP3 AGCGAATCAATGGACTCTGGAATATCC GTTTGCTGCATCGACATCTGTACCA CCL5 TCATTGCTACTGCCCTCTGCG ACTGCTGGGTTGGAGCACTTG CCL18 CCCTCCTTGTCCTCGTCTGCA GCACTGGGGGCTGGTTTCAG CCL26 TTCCAATACAGCCACAAGCCCC GGATGGGTACAGACTTTCTTGCCTC CCND2 TCAAGTGCGTGCAGAAGGACAT CTTCGCACTTCTGTTCCTCACA CCND3 TGGCTGCTGTGATTGCACATGA GATGGCGGGTACATGGCAAAGG CCR3 ACGCTGCTCTGCTTCCTGG TCCTCAGTTCCCCACCATCGC CCR4 AGCATCGTGCTTCCTGAGCAA GGTGTCTGCTATATCCGTGGGGT CCR7 AGACAGGGGTAGTGCGAGGC -

Robles JTO Supplemental Digital Content 1
Supplementary Materials An Integrated Prognostic Classifier for Stage I Lung Adenocarcinoma based on mRNA, microRNA and DNA Methylation Biomarkers Ana I. Robles1, Eri Arai2, Ewy A. Mathé1, Hirokazu Okayama1, Aaron Schetter1, Derek Brown1, David Petersen3, Elise D. Bowman1, Rintaro Noro1, Judith A. Welsh1, Daniel C. Edelman3, Holly S. Stevenson3, Yonghong Wang3, Naoto Tsuchiya4, Takashi Kohno4, Vidar Skaug5, Steen Mollerup5, Aage Haugen5, Paul S. Meltzer3, Jun Yokota6, Yae Kanai2 and Curtis C. Harris1 Affiliations: 1Laboratory of Human Carcinogenesis, NCI-CCR, National Institutes of Health, Bethesda, MD 20892, USA. 2Division of Molecular Pathology, National Cancer Center Research Institute, Tokyo 104-0045, Japan. 3Genetics Branch, NCI-CCR, National Institutes of Health, Bethesda, MD 20892, USA. 4Division of Genome Biology, National Cancer Center Research Institute, Tokyo 104-0045, Japan. 5Department of Chemical and Biological Working Environment, National Institute of Occupational Health, NO-0033 Oslo, Norway. 6Genomics and Epigenomics of Cancer Prediction Program, Institute of Predictive and Personalized Medicine of Cancer (IMPPC), 08916 Badalona (Barcelona), Spain. List of Supplementary Materials Supplementary Materials and Methods Fig. S1. Hierarchical clustering of based on CpG sites differentially-methylated in Stage I ADC compared to non-tumor adjacent tissues. Fig. S2. Confirmatory pyrosequencing analysis of DNA methylation at the HOXA9 locus in Stage I ADC from a subset of the NCI microarray cohort. 1 Fig. S3. Methylation Beta-values for HOXA9 probe cg26521404 in Stage I ADC samples from Japan. Fig. S4. Kaplan-Meier analysis of HOXA9 promoter methylation in a published cohort of Stage I lung ADC (J Clin Oncol 2013;31(32):4140-7). Fig. S5. Kaplan-Meier analysis of a combined prognostic biomarker in Stage I lung ADC. -

Meta-Analysis of Transcriptomic Variation in T Cell Populations Reveals Novel Signatures of Gene Expression and Splicing
bioRxiv preprint doi: https://doi.org/10.1101/727362; this version posted August 9, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. Meta-Analysis of Transcriptomic Variation in T cell Populations Reveals Novel Signatures of Gene Expression and Splicing Caleb M. Radens1, Davia Blake2, Paul Jewell4,5, Yoseph Barash1,4,5* and Kristen W. Lynch1,2,3,4*# 1 Cell and Molecular Biology and 2Immunology Graduate Groups and Departments of 3Biochemistry and Biophysics and 4Genetics, Perelman School of Medicine and 5Department of Computer Science, School of Engineering and Applied Science, University of Pennsylvania, Philadelphia, PA 19104, *Co-corresponding authors K.W.L.: [email protected] Y.B.: [email protected] #Lead Contact (K.W.L.: [email protected]) Running title: Gene Expression Signatures in T cell Subsets bioRxiv preprint doi: https://doi.org/10.1101/727362; this version posted August 9, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. Summary (150 words) Distinct T cell subtypes are typically defined by the expression of distinct gene repertoires. However, there is variability between studies regarding the markers used to define each T cell subtype. Moreover, previous analysis of gene expression in T cell subsets has largely focused on gene expression rather than alternative splicing. -

Antibody Repertoire and Gene Expression Dynamics of Diverse Human B Cell
bioRxiv preprint doi: https://doi.org/10.1101/2020.04.28.054775; this version posted May 26, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. 1 Antibody repertoire and gene expression dynamics of diverse human B cell 2 states during affinity maturation. 3 Hamish W King1,2 *, Nara Orban3, John C Riches4,5, Andrew J Clear4, Gary Warnes6, Sarah A 4 Teichmann2,7, Louisa K James1 * § 5 1 Centre for Immunobiology, Blizard Institute, Queen Mary University of London, London E1 2AT, UK 6 2 Wellcome Sanger Institute, Wellcome Genome Campus, Hinxton, Cambridge CB10 1SA, UK 7 3 Barts Health Ear, Nose and Throat Service, The Royal London Hospital, London E1 1BB, UK 8 4 Centre for Haemato-Oncology, Barts Cancer Institute, Queen Mary University of London, London EC1M 6BQ, UK 9 5 The Francis Crick Institute, London NW1 1AT, UK 10 6 Flow Cytometry Core Facility, Blizard Institute, Queen Mary University of London, London E1 2AT, UK 11 7 Theory of Condensed Matter, Cavendish Laboratory, Department of Physics, University of Cambridge, Cambridge CB3 0EH, UK 12 § Lead contact 13 * To whom correspondence can be addressed: [email protected], [email protected] and [email protected] 14 15 Abstract 16 In response to antigen challenge, B cells clonally expand, undergo selection and differentiate to produce 17 mature B cell subsets and high affinity antibodies.