Trachemys) Revealed by Coalescent Analyses of Mitochondrial and Cloned Nuclear Markers ⇑ James F
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
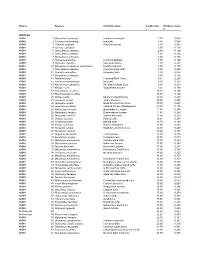
PDF File Containing Table of Lengths and Thicknesses of Turtle Shells And
Source Species Common name length (cm) thickness (cm) L t TURTLES AMNH 1 Sternotherus odoratus common musk turtle 2.30 0.089 AMNH 2 Clemmys muhlenbergi bug turtle 3.80 0.069 AMNH 3 Chersina angulata Angulate tortoise 3.90 0.050 AMNH 4 Testudo carbonera 6.97 0.130 AMNH 5 Sternotherus oderatus 6.99 0.160 AMNH 6 Sternotherus oderatus 7.00 0.165 AMNH 7 Sternotherus oderatus 7.00 0.165 AMNH 8 Homopus areolatus Common padloper 7.95 0.100 AMNH 9 Homopus signatus Speckled tortoise 7.98 0.231 AMNH 10 Kinosternon subrabum steinochneri Florida mud turtle 8.90 0.178 AMNH 11 Sternotherus oderatus Common musk turtle 8.98 0.290 AMNH 12 Chelydra serpentina Snapping turtle 8.98 0.076 AMNH 13 Sternotherus oderatus 9.00 0.168 AMNH 14 Hardella thurgi Crowned River Turtle 9.04 0.263 AMNH 15 Clemmys muhlenbergii Bog turtle 9.09 0.231 AMNH 16 Kinosternon subrubrum The Eastern Mud Turtle 9.10 0.253 AMNH 17 Kinixys crosa hinged-back tortoise 9.34 0.160 AMNH 18 Peamobates oculifers 10.17 0.140 AMNH 19 Peammobates oculifera 10.27 0.140 AMNH 20 Kinixys spekii Speke's hinged tortoise 10.30 0.201 AMNH 21 Terrapene ornata ornate box turtle 10.30 0.406 AMNH 22 Terrapene ornata North American box turtle 10.76 0.257 AMNH 23 Geochelone radiata radiated tortoise (Madagascar) 10.80 0.155 AMNH 24 Malaclemys terrapin diamondback terrapin 11.40 0.295 AMNH 25 Malaclemys terrapin Diamondback terrapin 11.58 0.264 AMNH 26 Terrapene carolina eastern box turtle 11.80 0.259 AMNH 27 Chrysemys picta Painted turtle 12.21 0.267 AMNH 28 Chrysemys picta painted turtle 12.70 0.168 AMNH 29 -

TCF Summary Activity Report 2002–2018
Turtle Conservation Fund • Summary Activity Report 2002–2018 Turtle Conservation Fund A Partnership Coalition of Leading Turtle Conservation Organizations and Individuals Summary Activity Report 2002–2018 1 Turtle Conservation Fund • Summary Activity Report 2002–2018 Recommended Citation: Turtle Conservation Fund [Rhodin, A.G.J., Quinn, H.R., Goode, E.V., Hudson, R., Mittermeier, R.A., and van Dijk, P.P.]. 2019. Turtle Conservation Fund: A Partnership Coalition of Leading Turtle Conservation Organi- zations and Individuals—Summary Activity Report 2002–2018. Lunenburg, MA and Ojai, CA: Chelonian Research Foundation and Turtle Conservancy, 54 pp. Front Cover Photo: Radiated Tortoise, Astrochelys radiata, Cap Sainte Marie Special Reserve, southern Madagascar. Photo by Anders G.J. Rhodin. Back Cover Photo: Yangtze Giant Softshell Turtle, Rafetus swinhoei, Dong Mo Lake, Hanoi, Vietnam. Photo by Timothy E.M. McCormack. Printed by Inkspot Press, Bennington, VT 05201 USA. Hardcopy available from Chelonian Research Foundation, 564 Chittenden Dr., Arlington, VT 05250 USA. Downloadable pdf copy available at www.turtleconservationfund.org 2 Turtle Conservation Fund • Summary Activity Report 2002–2018 Turtle Conservation Fund A Partnership Coalition of Leading Turtle Conservation Organizations and Individuals Summary Activity Report 2002–2018 by Anders G.J. Rhodin, Hugh R. Quinn, Eric V. Goode, Rick Hudson, Russell A. Mittermeier, and Peter Paul van Dijk Strategic Action Planning and Funding Support for Conservation of Threatened Tortoises and Freshwater -

Cfreptiles & Amphibians
WWW.IRCF.ORG/REPTILESANDAMPHIBIANSJOURNALTABLE OF CONTENTS IRCF REPTILES & IRCF AMPHIBIANS REPTILES • VOL &15, AMPHIBIANS NO 4 • DEC 2008 • 19(2):143–147189 • JUNE 2012 IRCF REPTILES & AMPHIBIANS CONSERVATION AND NATURAL HISTORY TABLE OF CONTENTS NEWSBRIEFS FEATURE ARTICLES . Chasing Bullsnakes (Pituophis catenifer sayi) in Wisconsin: A GrowingOn the Road Illicit to Understanding Trade the Ecology and Conservation of the Midwest’s Giant Serpent ...................... Joshua M. Kapfer 190 . The Shared History of Treeboas (Corallus grenadensis) and Humans on Grenada: 1 Threatens AJamaica’s Hypothetical Excursion Wildlife ............................................................................................................................Robert W. Henderson 198 Conservationists areRESEARCH beginning ARTICLESto fear that as demand grows for rare and exotic .wildlife,The Texas Horned Jamaican Lizard in Central authorities and Western Texaswill ....................... be Emily Henry, Jason Brewer, Krista Mougey, and Gad Perry 204 unable to protect the. island’sThe Knight naturalAnole (Anolis treasures. equestris) in Florida Local wildlife .............................................Brian J. Camposano, Kenneth L. Krysko, Kevin M. Enge, Ellen M. Donlan, and Michael Granatosky 212 is commonly offered for sale to tourists across the island and the National EnvironmentCONSERVATION and Planning ALERT Agency (NEPA), . World’s Mammals in Crisis ............................................................................................................................................................ -

A Systematic Review of the Turtle Family Emydidae
67 (1): 1 – 122 © Senckenberg Gesellschaft für Naturforschung, 2017. 30.6.2017 A Systematic Review of the Turtle Family Emydidae Michael E. Seidel1 & Carl H. Ernst 2 1 4430 Richmond Park Drive East, Jacksonville, FL, 32224, USA and Department of Biological Sciences, Marshall University, Huntington, WV, USA; [email protected] — 2 Division of Amphibians and Reptiles, mrc 162, Smithsonian Institution, P.O. Box 37012, Washington, D.C. 200137012, USA; [email protected] Accepted 19.ix.2016. Published online at www.senckenberg.de / vertebrate-zoology on 27.vi.2016. Abstract Family Emydidae is a large and diverse group of turtles comprised of 50 – 60 extant species. After a long history of taxonomic revision, the family is presently recognized as a monophyletic group defined by unique skeletal and molecular character states. Emydids are believed to have originated in the Eocene, 42 – 56 million years ago. They are mostly native to North America, but one genus, Trachemys, occurs in South America and a second, Emys, ranges over parts of Europe, western Asia, and northern Africa. Some of the species are threatened and their future survival depends in part on understanding their systematic relationships and habitat requirements. The present treatise provides a synthesis and update of studies which define diversity and classification of the Emydidae. A review of family nomenclature indicates that RAFINESQUE, 1815 should be credited for the family name Emydidae. Early taxonomic studies of these turtles were based primarily on morphological data, including some fossil material. More recent work has relied heavily on phylogenetic analyses using molecular data, mostly DNA. The bulk of current evidence supports two major lineages: the subfamily Emydinae which has mostly semi-terrestrial forms ( genera Actinemys, Clemmys, Emydoidea, Emys, Glyptemys, Terrapene) and the more aquatic subfamily Deirochelyinae ( genera Chrysemys, Deirochelys, Graptemys, Malaclemys, Pseudemys, Trachemys). -

Chelonian Advisory Group Regional Collection Plan 4Th Edition December 2015
Association of Zoos and Aquariums (AZA) Chelonian Advisory Group Regional Collection Plan 4th Edition December 2015 Editor Chelonian TAG Steering Committee 1 TABLE OF CONTENTS Introduction Mission ...................................................................................................................................... 3 Steering Committee Structure ........................................................................................................... 3 Officers, Steering Committee Members, and Advisors ..................................................................... 4 Taxonomic Scope ............................................................................................................................. 6 Space Analysis Space .......................................................................................................................................... 6 Survey ........................................................................................................................................ 6 Current and Potential Holding Table Results ............................................................................. 8 Species Selection Process Process ..................................................................................................................................... 11 Decision Tree ........................................................................................................................... 13 Decision Tree Results ............................................................................................................. -

Turtles of the World, 2010 Update: Annotated Checklist of Taxonomy, Synonymy, Distribution, and Conservation Status
Conservation Biology of Freshwater Turtles and Tortoises: A Compilation ProjectTurtles of the IUCN/SSC of the World Tortoise – 2010and Freshwater Checklist Turtle Specialist Group 000.85 A.G.J. Rhodin, P.C.H. Pritchard, P.P. van Dijk, R.A. Saumure, K.A. Buhlmann, J.B. Iverson, and R.A. Mittermeier, Eds. Chelonian Research Monographs (ISSN 1088-7105) No. 5, doi:10.3854/crm.5.000.checklist.v3.2010 © 2010 by Chelonian Research Foundation • Published 14 December 2010 Turtles of the World, 2010 Update: Annotated Checklist of Taxonomy, Synonymy, Distribution, and Conservation Status TUR T LE TAXONOMY WORKING GROUP * *Authorship of this article is by this working group of the IUCN/SSC Tortoise and Freshwater Turtle Specialist Group, which for the purposes of this document consisted of the following contributors: ANDERS G.J. RHODIN 1, PE T ER PAUL VAN DI J K 2, JOHN B. IVERSON 3, AND H. BRADLEY SHAFFER 4 1Chair, IUCN/SSC Tortoise and Freshwater Turtle Specialist Group, Chelonian Research Foundation, 168 Goodrich St., Lunenburg, Massachusetts 01462 USA [[email protected]]; 2Deputy Chair, IUCN/SSC Tortoise and Freshwater Turtle Specialist Group, Conservation International, 2011 Crystal Drive, Suite 500, Arlington, Virginia 22202 USA [[email protected]]; 3Department of Biology, Earlham College, Richmond, Indiana 47374 USA [[email protected]]; 4Department of Evolution and Ecology, University of California, Davis, California 95616 USA [[email protected]] AB S T RAC T . – This is our fourth annual compilation of an annotated checklist of all recognized and named taxa of the world’s modern chelonian fauna, documenting recent changes and controversies in nomenclature, and including all primary synonyms, updated from our previous three checklists (Turtle Taxonomy Working Group [2007b, 2009], Rhodin et al. -

Norntates PUBLISHED by the AMERICAN MUSEUM of NATURAL HISTORY CENTRAL PARK WEST at 79TH STREET, NEW YORK, N.Y
AMERICANt MUSEUM Norntates PUBLISHED BY THE AMERICAN MUSEUM OF NATURAL HISTORY CENTRAL PARK WEST AT 79TH STREET, NEW YORK, N.Y. 10024 Number 2918, pp. 1-41, figs. 1-24, tables 1-3 June 28, 1988 Revision of the West Indian Emydid Turtles (Testudines) MICHAEL E. SEIDEL' ABSTRACT The systematics ofemydid turtles, genus Trach- granti (Barbour and Carr), T. d. plana (Barbour emys, in the West Indies is analyzed using external and Carr)-are placed in the synonymies of T. morphology, osteology, and biochemical data. Re- terrapen, T. d. angusta, and T. d. decussata, re- sults indicate the presence of seven recognizable spectively. The description of each valid taxon taxa: T. terrapen in Jamaica and the northern Ba- includes a complete synonymy, new diagnostic hamas, T. decorata on Hispaniola, T. stejnegeri characters, distribution, and ecological notes. Also stejnegeri in Puerto Rico, T. s. vicina on Hispan- included are a taxonomic key and discussion of iola, T. s. malonei on Great Inagua (southern Ba- possible evolutionary origins. Phylogenetic evi- hamas), T. decussata decussata in Cuba, and T. d. dence suggests that the West Indian segment of angusta in Cuba and the Cayman Islands. Three Trachemys is paraphyletic and therefore does not previously described taxa-T. felis (Barbour), T. constitute a natural taxonomic complex. RESUMEN La taxonomia de las tortugas emydid del genero d. plana (Barbour y Carr)-estan puestas en si- Trachemys de Las Indias Occidentales esta analy- nonimias de T. terrapen, T. d. angusta y T. d. zada usando morfologia externa, osteologia, y da- decussata, respectivamente. La descripcion de cada tos bioquimicos. -

On the Brink of Extinction: Saving Jamaica’S Vanishing Species
The Environmental Foundation of Jamaica in association with Jamaica Conservation and Development Trust present The 7th Annual EFJ Public Lecture On the Brink of Extinction: Saving Jamaica’s Vanishing Species Dr. Byron Wilson, Senior Lecturer, Department of Life Sciences, University of the West Indies, Mona October 20, 2011, 5:30 pm FOREWORD The Environmental Foundation of Jamaica (EFJ) is very pleased to welcome you to our 7th Annual Public Lecture. The Lecture this year is being organized in partnership with our member and long standing partners at the Jamaica Conservation and Development Trust (JCDT). JCDT has been a major player not only in Protected Areas and Management of the Blue and John Crow Mountain National Park, but also in the area of public awareness and education with features such as the National Green Expo held every other year. Like all our work over the last 18 years, we have balanced the Public Lectures between our two major areas of focus – Child Development and Environmental Management. This year, our environmental lecture will focus on issues of biodiversity and human impact on ecological habitats. Our lecturer, Dr. Byron Wilson is a Senior Lecturer in the University of the West Indies. He is an expert in conservation biology and has dedicated his research to the ecology and conservation of the critically endangered Jamaican Iguana and other rare endemic reptile species. In addition to Dr. Wilson’s work, the EFJ has partnered with Government, Researchers and Conservationists to study and record critical information and data on Endemic and Endangered Species across Jamaica, including the establishment of the UWI/EFJ Biodiversity Centre in Port Royal, the Virtual Herbarium and the publication of the “Endemic Trees of Jamaica” book. -

Biolphilately Vol-64 No-1
Vol. 64 (1) Biophilately March 2015 41 HERPETOLOGY Temporary Editor Jack R. Congrove, BU1424 [Ed. Note: I am still looking for someone to take over as Associate Editor for this column. This listing is longer than usual because it compensates for the omission in the past two editions and it also includes a large quantity of entries that have not yet received catalog listing. It is doubtful that Scott will ever list some of these entries because they may have been produced without official sanction by the postal authorities of the country they purport to be from. Nonetheless you can find them in the marketplace, so caveat emptor.] Scott# Denom Common Name/Scientific Name Family/Subfamily Code ALBANIA 2007 October 17 (Arms Type of 2001) 2803 60L Stylized Snake wrapped around branch (Kokini coat of arms) S 2013 November 6 (17th Mediterranean Games) 2943a 40L Stylized Sea Turtle (games logo) S 2943b 150L Stylized Sea Turtle (games logo) S 2943 Horiz pair (Sc#2943a–b) 2944 SS 200L LL: Stylized Sea Turtle (games logo) S Z ARGENTINA 2013 June 1 (50th anniv. African Union) 2683 4p Stylized Lizard silhouette in outline of African continent S ARUBA 2013 April 5 (Currency) 425d 200c Stylized Frog on 100 Florin note S 2013 November 12 (Nature Photography) 432c 120c Cope’s Ameiva, Ameiva bifrontata Teiidae A* 432 Block of 10 (Sc#432a–j) 2013 December 16 (Wildlife) 433a 175c Red-eyed Tree Frog, Agalychnis callidryas (Cap: A. callidryas) Hylidae A* 433d 175c Blessed Poison Frog, Ranitomeya benedicta (Cap: Ranitomega) Dendrobatidae A* 433 Block of 8 (Sc#433a–h) -

Ecology of the Jamaican Slider Turtle (Trachemys Terrapen), with Implications for Conservation and Management
Ch<"lo11iun Co,tSt'n'CJtion <md Biology, 2005. -1(4):908-915 0 2005 by CheJonian Research Foundation Ecology of the Jamaican Slider Turtle (Trachemys terrapen), with Implications for Conservation and Management TRACEY D. T UBERVILLE'' K URT A. BUHLMANN'- 2, RHEMA KERR BJORKL AND3, AND Do uG BooHER 3 1Savc11111ahRiver Ecology laborato,y, P.O. Drawer£, Aiken, South Carolina 29802 USA f Fax: 803-725-3309: £-mails: [email protected]; [email protected]/: 2Co11servatio11l11t ema tio11al.Centerfor Applied Biodiversity Science, 1919 M Street NW, Washington, D.C. 20036 USA: 'University of Georgia, Institute of Ecology. Athens. Georgia 30602 USA {E-mail: [email protected]] AnsTRACJ', - We investigated population s of the Jamaican slider turtle (Trachemys terrapen), a species apparently endemic to Jamaica and the only native freshwater turtle species known to occur there. We captured 54 turtles at four sites (three along the southern coast and one in the northw est) representing a variety of habitats, includin g a permanentl y ponded wetland, farm ponds, and a stream in karst landscape. Turtles were also found in a series of seasonal ponds where they retreat into cave refugia during dry periods when caves remain nooded , thus allowing the slider population to exist in this seasonally arid landscape. We did not observe or capture turtles during limited sampling in a large river or a brackish mangrove swamp. Individuals from the northwest population (n = 12) were morpholo gically distinct from turtles captured along the south coast (11= 42 ) and descriptions provided in the literature for T. terrapen. Jamaican slider turtles are harvested incidentall y by local residents wherever they are found, and concentrated population s, such as those in cave refugia, are heavily exploited. -

ZOO 4462C – Herpetology Spring 2021, 4 Credits
ZOO 4462C – Herpetology Spring 2021, 4 credits Course Schedule – See page 10 Instructor: Dr. Gregg Klowden (pronounced "Cloud - in”) Office: Room 202A, Biological Sciences E-mail: [email protected] Phone: Please send an email instead Mark Catesby (1731) “Natural History of Carolina, Florida and the Bahama Islands” "These foul and loathsome animals . are abhorrent because of their cold body, pale color, cartilaginous skeleton, filthy skin, fierce aspect, calculating eye, offensive smell, harsh voice, squalid habitation, and terrible venom; and so their Creator has not exerted his powers to make many of them." Carolus Linnaeus (1758) ***Email Requirements: I teach several courses and receive a large volume of emails. To help me help you please: 1. format the subject of your email as follows: “Course – Herpetology, Subject - Question about exam 1” 2. include your 1st and last name in the body of all correspondence. I try to respond to emails within 48 hours however, response time may be greater. o Please plan accordingly by not waiting to the last minute to contact me with questions or concerns. All messaging must be done using either Webcourses or your Knight's E-Mail. o Messages from non-UCF addresses will not be answered. Due to confidentiality, questions about grades should be sent via Webcourses messaging, not via email. Office Hours: Tuesdays and Thursdays 10:30-11:30a and 2:00-3:00p or by appointment All office hours will be held online via Zoom. An appointment is not necessary. Just log into Zoom using the link posted on Webcourses. You will initially be admitted to a waiting room and Dr. -
Biologia E Conservação De Quelônios No Delta Do Rio Jacuí – Rs: Aspectos Da História Natural De Espécies Em Ambientes Alterados Pelo Homem
CLÓVIS DE SOUZA BUJES BIOLOGIA E CONSERVAÇÃO DE QUELÔNIOS NO DELTA DO RIO JACUÍ – RS: ASPECTOS DA HISTÓRIA NATURAL DE ESPÉCIES EM AMBIENTES ALTERADOS PELO HOMEM Tese de Doutorado apresentada ao Programa de Pós-Graduação em Biologia Animal, Instituto de Biociências, da Universidade Federal do Rio Grande do Sul, como requisito parcial à obtenção do título de Doutor em Biologia Animal. Área de concentração: Biodiversidade Orientadora: Profa. Dra. Laura Verrastro Viñas UNIVERSIDADE FEDERAL DO RIO GRANDE DO SUL INSTITUTO DE BIOCIÊNCIAS PORTO ALEGRE 2008 ao meU aMor, a miNha vidA, a quEm me enTende, da foRma Como sOu, na mInhA essência, nos meUs exceSsos e, principaLmentE, na maneiRa cOmo veJo a vida. iii AGRADECIMENTOS A Dra. Laura Verrastro Viñas pela orientação, apoio, incentivo e amizade. Aos membros da banca de acompanhamento de doutorado, Dra. Marta Elena Fabian, Dra. Clarice Bernhardt Fialho, Dr. Thales Renato Ochotorena de Freitas e Dr. Márcio Borges Martins. Ao Programa de Pós-Graduação em Biologia Animal e a seus coordenadores, Dr. Luiz Roberto Malabarba e Dra. Clarice Bernhardt Fialho. A Fundação O Boticário de Proteção à Natureza (FBPN) pelos dois anos de apoio financeiro ao Projeto Chelonia–RS (Processo 0594-20032). Ao Instituto Gaúcho de Estudos Ambientais (INGA), à Secretaria Estadual do Meio Ambiente (SEMA) e ao Departamento de Zoologia da Universidade Federal do Rio Grande do Sul (UFRGS) pelo apoio logístico. A Loiva, ao Clemente e aos demais funcionários do Parque Estadual Delta do Jacuí por suas receptividade e colaboração. Aos membros da banca examinadora por suas críticas e sugestões. A todos os colegas do laboratório de Herpetologia que, direta ou indiretamente, contribuíram para o enriquecimento deste trabalho e, especialmente, a Priscila Miorando, a Bettina Marks, a Isabel Ely, a Renata Cardoso e a Júlia Witt por sua colaboração nas tarefas de campo, de laboratório e de educação ambiental.