Protein Interactions Allow Functional Regulation of Homocysteine Metabolism
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Propranolol-Mediated Attenuation of MMP-9 Excretion in Infants with Hemangiomas
Supplementary Online Content Thaivalappil S, Bauman N, Saieg A, Movius E, Brown KJ, Preciado D. Propranolol-mediated attenuation of MMP-9 excretion in infants with hemangiomas. JAMA Otolaryngol Head Neck Surg. doi:10.1001/jamaoto.2013.4773 eTable. List of All of the Proteins Identified by Proteomics This supplementary material has been provided by the authors to give readers additional information about their work. © 2013 American Medical Association. All rights reserved. Downloaded From: https://jamanetwork.com/ on 10/01/2021 eTable. List of All of the Proteins Identified by Proteomics Protein Name Prop 12 mo/4 Pred 12 mo/4 Δ Prop to Pred mo mo Myeloperoxidase OS=Homo sapiens GN=MPO 26.00 143.00 ‐117.00 Lactotransferrin OS=Homo sapiens GN=LTF 114.00 205.50 ‐91.50 Matrix metalloproteinase‐9 OS=Homo sapiens GN=MMP9 5.00 36.00 ‐31.00 Neutrophil elastase OS=Homo sapiens GN=ELANE 24.00 48.00 ‐24.00 Bleomycin hydrolase OS=Homo sapiens GN=BLMH 3.00 25.00 ‐22.00 CAP7_HUMAN Azurocidin OS=Homo sapiens GN=AZU1 PE=1 SV=3 4.00 26.00 ‐22.00 S10A8_HUMAN Protein S100‐A8 OS=Homo sapiens GN=S100A8 PE=1 14.67 30.50 ‐15.83 SV=1 IL1F9_HUMAN Interleukin‐1 family member 9 OS=Homo sapiens 1.00 15.00 ‐14.00 GN=IL1F9 PE=1 SV=1 MUC5B_HUMAN Mucin‐5B OS=Homo sapiens GN=MUC5B PE=1 SV=3 2.00 14.00 ‐12.00 MUC4_HUMAN Mucin‐4 OS=Homo sapiens GN=MUC4 PE=1 SV=3 1.00 12.00 ‐11.00 HRG_HUMAN Histidine‐rich glycoprotein OS=Homo sapiens GN=HRG 1.00 12.00 ‐11.00 PE=1 SV=1 TKT_HUMAN Transketolase OS=Homo sapiens GN=TKT PE=1 SV=3 17.00 28.00 ‐11.00 CATG_HUMAN Cathepsin G OS=Homo -

Exosomes Confer Chemoresistance to Pancreatic Cancer Cells By
FULL PAPER British Journal of Cancer (2017) 116, 609–619 | doi: 10.1038/bjc.2017.18 Keywords: chemoresistance; exosomes; pancreatic cancer; ROS; microRNA Exosomes confer chemoresistance to pancreatic cancer cells by promoting ROS detoxification and miR-155-mediated suppression of key gemcitabine-metabolising enzyme, DCK Girijesh Kumar Patel1, Mohammad Aslam Khan1, Arun Bhardwaj1, Sanjeev K Srivastava1, Haseeb Zubair1, Mary C Patton1, Seema Singh1,2, Moh’d Khushman3 and Ajay P Singh*,1,2 1Department of Oncologic Sciences, Mitchell Cancer Institute, University of South Alabama, Mobile, AL, USA; 2Department of Biochemistry and Molecular Biology, College of Medicine, University of South Alabama, Mobile, AL, USA and 3Department of Interdisciplinary Clinical Oncology, Mitchell Cancer Institute, University of South Alabama, Mobile, AL, USA Background: Chemoresistance is a significant clinical problem in pancreatic cancer (PC) and underlying molecular mechanisms still remain to be completely understood. Here we report a novel exosome-mediated mechanism of drug-induced acquired chemoresistance in PC cells. Methods: Differential ultracentrifugation was performed to isolate extracellular vesicles (EVs) based on their size from vehicle- or gemcitabine-treated PC cells. Extracellular vesicles size and subtypes were determined by dynamic light scattering and marker profiling, respectively. Gene expression was examined by qRT-PCR and/or immunoblot analyses, and direct targeting of DCK by miR-155 was confirmed by dual-luciferase 30-UTR reporter assay. Flow cytometry was performed to examine the apoptosis indices and reactive oxygen species (ROS) levels in PC cells using specific dyes. Cell viability was determined using the WST-1 assay. Results: Conditioned media (CM) from gemcitabine-treated PC cells (Gem-CM) provided significant chemoprotection to subsequent gemcitabine toxicity and most of the chemoresistance conferred by Gem-CM resulted from its EVs fraction. -

Pseudouridine Synthase 1: a Site-Specific Synthase Without Strict Sequence Recognition Requirements Bryan S
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by PubMed Central Published online 18 November 2011 Nucleic Acids Research, 2012, Vol. 40, No. 5 2107–2118 doi:10.1093/nar/gkr1017 Pseudouridine synthase 1: a site-specific synthase without strict sequence recognition requirements Bryan S. Sibert and Jeffrey R. Patton* Department of Pathology, Microbiology and Immunology, University of South Carolina, School of Medicine, Columbia, SC 29208 USA Received May 20, 2011; Revised October 19, 2011; Accepted October 22, 2011 ABSTRACT rRNA and snRNA and requires Dyskerin or its homologs Pseudouridine synthase 1 (Pus1p) is an unusual (Cbf5p in yeast for example) and RNP cofactors [most site-specific modification enzyme in that it can often H/ACA small nucleolar ribonucleoprotein particles modify a number of positions in tRNAs and can rec- (snoRNPs)] that enable one enzyme to recognize many ognize several other types of RNA. No consensus different sites for modification on different substrates recognition sequence or structure has been identi- (17–25). The other pathway for É formation employs fied for Pus1p. Human Pus1p was used to determine site-specific É synthases that require no cofactors to rec- which structural or sequence elements of human ognize and modify the RNA substrate. A number of en- tRNASer are necessary for pseudouridine ()) forma- zymes have been identified in this pathway and are grouped in six families that all share a common basic tion at position 28 in the anticodon stem-loop (ASL). Ser structure (4). It is safe to say the cofactor ‘guided’ pathway Some point mutations in the ASL stem of tRNA has received a great deal of attention because of its simi- had significant effects on the levels of modification larity to aspects of RNA editing, but the site-specific and compensatory mutation, to reform the base pseudouridine synthases accomplish the same task, on pair, restored a wild-type level of ) formation. -

Identification of Transcriptomic Differences Between Lower
International Journal of Molecular Sciences Article Identification of Transcriptomic Differences between Lower Extremities Arterial Disease, Abdominal Aortic Aneurysm and Chronic Venous Disease in Peripheral Blood Mononuclear Cells Specimens Daniel P. Zalewski 1,*,† , Karol P. Ruszel 2,†, Andrzej St˛epniewski 3, Dariusz Gałkowski 4, Jacek Bogucki 5 , Przemysław Kołodziej 6 , Jolanta Szyma ´nska 7 , Bartosz J. Płachno 8 , Tomasz Zubilewicz 9 , Marcin Feldo 9,‡ , Janusz Kocki 2,‡ and Anna Bogucka-Kocka 1,‡ 1 Chair and Department of Biology and Genetics, Medical University of Lublin, 4a Chod´zkiSt., 20-093 Lublin, Poland; [email protected] 2 Chair of Medical Genetics, Department of Clinical Genetics, Medical University of Lublin, 11 Radziwiłłowska St., 20-080 Lublin, Poland; [email protected] (K.P.R.); [email protected] (J.K.) 3 Ecotech Complex Analytical and Programme Centre for Advanced Environmentally Friendly Technologies, University of Marie Curie-Skłodowska, 39 Gł˛ebokaSt., 20-612 Lublin, Poland; [email protected] 4 Department of Pathology and Laboratory Medicine, Rutgers-Robert Wood Johnson Medical School, One Robert Wood Johnson Place, New Brunswick, NJ 08903-0019, USA; [email protected] 5 Chair and Department of Organic Chemistry, Medical University of Lublin, 4a Chod´zkiSt., Citation: Zalewski, D.P.; Ruszel, K.P.; 20-093 Lublin, Poland; [email protected] St˛epniewski,A.; Gałkowski, D.; 6 Laboratory of Diagnostic Parasitology, Chair and Department of Biology and Genetics, Medical University of Bogucki, J.; Kołodziej, P.; Szyma´nska, Lublin, 4a Chod´zkiSt., 20-093 Lublin, Poland; [email protected] J.; Płachno, B.J.; Zubilewicz, T.; Feldo, 7 Department of Integrated Paediatric Dentistry, Chair of Integrated Dentistry, Medical University of Lublin, M.; et al. -

TITLE PAGE Oxidative Stress and Response to Thymidylate Synthase
Downloaded from molpharm.aspetjournals.org at ASPET Journals on October 2, 2021 -Targeted -Targeted 1 , University of of , University SC K.W.B., South Columbia, (U.O., Carolina, This article has not been copyedited and formatted. The final version may differ from this version. This article has not been copyedited and formatted. The final version may differ from this version. This article has not been copyedited and formatted. The final version may differ from this version. This article has not been copyedited and formatted. The final version may differ from this version. This article has not been copyedited and formatted. The final version may differ from this version. This article has not been copyedited and formatted. The final version may differ from this version. This article has not been copyedited and formatted. The final version may differ from this version. This article has not been copyedited and formatted. The final version may differ from this version. This article has not been copyedited and formatted. The final version may differ from this version. This article has not been copyedited and formatted. The final version may differ from this version. This article has not been copyedited and formatted. The final version may differ from this version. This article has not been copyedited and formatted. The final version may differ from this version. This article has not been copyedited and formatted. The final version may differ from this version. This article has not been copyedited and formatted. The final version may differ from this version. This article has not been copyedited and formatted. -

Supplementary Table S4. FGA Co-Expressed Gene List in LUAD
Supplementary Table S4. FGA co-expressed gene list in LUAD tumors Symbol R Locus Description FGG 0.919 4q28 fibrinogen gamma chain FGL1 0.635 8p22 fibrinogen-like 1 SLC7A2 0.536 8p22 solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 DUSP4 0.521 8p12-p11 dual specificity phosphatase 4 HAL 0.51 12q22-q24.1histidine ammonia-lyase PDE4D 0.499 5q12 phosphodiesterase 4D, cAMP-specific FURIN 0.497 15q26.1 furin (paired basic amino acid cleaving enzyme) CPS1 0.49 2q35 carbamoyl-phosphate synthase 1, mitochondrial TESC 0.478 12q24.22 tescalcin INHA 0.465 2q35 inhibin, alpha S100P 0.461 4p16 S100 calcium binding protein P VPS37A 0.447 8p22 vacuolar protein sorting 37 homolog A (S. cerevisiae) SLC16A14 0.447 2q36.3 solute carrier family 16, member 14 PPARGC1A 0.443 4p15.1 peroxisome proliferator-activated receptor gamma, coactivator 1 alpha SIK1 0.435 21q22.3 salt-inducible kinase 1 IRS2 0.434 13q34 insulin receptor substrate 2 RND1 0.433 12q12 Rho family GTPase 1 HGD 0.433 3q13.33 homogentisate 1,2-dioxygenase PTP4A1 0.432 6q12 protein tyrosine phosphatase type IVA, member 1 C8orf4 0.428 8p11.2 chromosome 8 open reading frame 4 DDC 0.427 7p12.2 dopa decarboxylase (aromatic L-amino acid decarboxylase) TACC2 0.427 10q26 transforming, acidic coiled-coil containing protein 2 MUC13 0.422 3q21.2 mucin 13, cell surface associated C5 0.412 9q33-q34 complement component 5 NR4A2 0.412 2q22-q23 nuclear receptor subfamily 4, group A, member 2 EYS 0.411 6q12 eyes shut homolog (Drosophila) GPX2 0.406 14q24.1 glutathione peroxidase -

Computational Modeling of Lysine Post-Translational Modification: an Overview Md
c and S eti ys h te nt m y s S B Hasan MM et al., Curr Synthetic Sys Biol 2018, 6:1 t i n o e l Current Synthetic and o r r g DOI: 10.4172/2332-0737.1000137 u y C ISSN: 2332-0737 Systems Biology CommentaryResearch Article OpenOpen Access Access Computational Modeling of Lysine Post-Translational Modification: An Overview Md. Mehedi Hasan 1*, Mst. Shamima Khatun2, and Hiroyuki Kurata1,3 1Department of Bioscience and Bioinformatics, Kyushu Institute of Technology, 680-4 Kawazu, Iizuka, Fukuoka 820-8502, Japan 2Department of Statistics, Laboratory of Bioinformatics, Rajshahi University-6205, Bangladesh 3Biomedical Informatics R&D Center, Kyushu Institute of Technology, 680-4 Kawazu, Iizuka, Fukuoka 820-8502, Japan Commentary hot spot for PTMs, and a number of protein lysine modifications could occur in both histone and non-histone proteins [11,12]. For instance, Living organisms have a magnificent ordered and complex lysine methylation in non-histone proteins can regulate the protein structure. In regulating the cellular functions, post-translational activity and protein structure stability [13]. In 2004, the Nobel Prize in modifications (PTMs) are critical molecular measures. They alter Chemistry was awarded jointly to Aaron Ciechanover, Avram Hershko protein conformation, modulating their activity, stability and and Irwin Rose for the discovery of lysine ubiquitin-mediated protein localization. Up to date, more than 300 types of PTMs are experimentally degradation [14]. discovered in vivo and in vitro pathways [1,2]. Major and common PTMs are methylation, ubiquitination, succinylation, phosphorylation, Moreover, in biological process, lysine can be modified by the glycosylation, acetylation, and sumoylation. -

Neuronal Network Dysfunction in a Human Model for Kleefstra Syndrome Mediated by Enhanced NMDAR Signaling
bioRxiv preprint doi: https://doi.org/10.1101/585596; this version posted March 21, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Neuronal network dysfunction in a human model for Kleefstra syndrome mediated by enhanced NMDAR signaling Monica Frega1,2†, Katrin Linda1†, Jason M. Keller1, Güvem Gümüş-Akay1, Britt Mossink1, Jon- Ruben van Rhijn3, Moritz Negwer1, Teun Klein Gunnewiek4, Katharina Foreman3, Nine Kompier3, Chantal Schoenmaker1, Willem van den Akker1, Astrid Oudakker1, Huiqing Zhou1,5, Tjitske Kleefstra1, Dirk Schubert3, Hans van Bokhoven1,3, Nael Nadif Kasri1,3* 1Department of Human Genetics, Radboudumc, Donders Institute for Brain, Cognition, and Behaviour, 6500 HB Nijmegen, the Netherlands 2Department of Clinical neurophysiology, University of Twente, 7522 NB Enschede, the Netherlands 3Department of Cognitive Neuroscience, Radboudumc, Donders Institute for Brain, Cognition and Behaviour, 6500 HB Nijmegen, the Netherlands 4Department of Anatomy, Radboudumc, Donders Institute for Brain, Cognition and Behaviour, 6500 HB Nijmegen, the Netherlands 5Department of Molecular Developmental Biology, Faculty of Science, Radboud Institute for Molecular Life Sciences, Radboud University, 6500 HB Nijmegen, the Netherlands †These authors contributed equally to the work *Corresponding Author dr. Nael Nadif Kasri Email: [email protected] bioRxiv preprint doi: https://doi.org/10.1101/585596; this version posted March 21, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Abstract Epigenetic regulation of gene transcription plays a critical role in neural network development and in the etiology of Intellectual Disability (ID) and Autism Spectrum Disorder (ASD). -
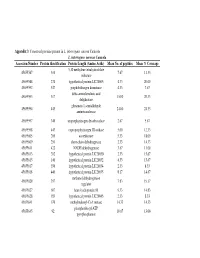
Appendix 3 and 4
Appendix 3 : Conserved proteins present in L. interrogans serovar Canicola L. interrogans serovar Canicola Accession Number Protein identification Protein Length (Amino Acids) Mean No. of peptides Mean % Coverage 5,10 methylene tetrahydrofolate 45655587 310 7.67 11.33 reductase 45655588 274 hypothetical protein LIC20005 4.33 20.00 45655592 547 porphobilinogen deaminase 4.33 7.67 delta-aminolevulinic acid 45655593 317 15.00 20.33 dehydratase glutamate-1-semialdehyde 45655594 443 24.00 24.33 aminotransferase 45655597 340 uroporphyrinogen decarboxylase 2.67 5.67 45655598 443 coproporphyrinogen III oxidase 5.00 12.33 45655605 200 azoreductase 5.33 18.00 45655609 251 short-chain dehydrogenase 2.33 14.33 45655611 422 NADH dehydrogenase 3.67 11.00 45655613 202 hypothetical protein LIC20030 2.33 15.67 45655615 140 hypothetical protein LIC20032 4.33 15.67 45655617 356 hypothetical protein LIC20034 2.33 8.33 45655618 440 hypothetical protein LIC20035 8.17 14.67 methanol dehydrogenase 45655620 357 7.83 19.17 regulator 45655627 607 heat shock protein 90 9.33 14.83 45655628 189 hypothetical protein LIC20045 2.33 8.33 45655641 670 methylmalonyl-CoA mutase 14.33 14.33 phosphoribosyl-ATP 45655645 92 10.67 13.00 pyrophosphatase 3-oxoacyl-(acyl-carrier protein) 45655647 254 4.00 17.00 reductase 45655648 77 acyl carrier protein 3.67 9.00 45655661 171 hypothetical protein LIC20078 5.00 26.00 4-hydroxybenzoyl-CoA 45655663 142 3.33 11.67 thioesterase S-adenosyl-L-homocysteine 45655666 436 13.00 19.67 hydrolase B12-dependent methionine 45655668 1247 42.67 21.67 -

Loss of Conserved Ubiquitylation Sites in Conserved Proteins During Human Evolution
INTERNATIONAL JOURNAL OF MOleCular meDICine 42: 2203-2212, 2018 Loss of conserved ubiquitylation sites in conserved proteins during human evolution DONGBIN PARK, CHUL JUN GOH, HYEIN KIM, JI SEOK LEE and YOONSOO HAHN Department of Life Science, Chung‑Ang University, Seoul 06974, Republic of Korea Received January 30, 2018; Accepted July 6, 2018 DOI: 10.3892/ijmm.2018.3772 Abstract. Ubiquitylation of lysine residues in proteins serves Introduction a pivotal role in the efficient removal of misfolded or unused proteins and in the control of various regulatory pathways Ubiquitylation, in which the highly conserved 76‑residue poly- by monitoring protein activity that may lead to protein peptide ubiquitin is covalently attached to a lysine residue of degradation. The loss of ubiquitylated lysines may affect substrate proteins, mediates the targeted destruction of ubiq- the ubiquitin‑mediated regulatory network and result in the uitylated proteins by the ubiquitin‑proteasome system (1‑4). emergence of novel phenotypes. The present study analyzed The ubiquitin‑mediated protein degradation pathway serves a mouse ubiquitylation data and orthologous proteins from crucial role in the efficient and specific removal of misfolded 62 mammals to identify 193 conserved ubiquitylation sites from proteins and certain key regulatory proteins (5,6). Ubiquitin 169 proteins that were lost in the Euarchonta lineage leading and other ubiquitin‑like proteins, including autophagy‑related to humans. A total of 8 proteins, including betaine homo- protein 8, Ubiquitin‑like -

Knockdown of RRM1 with Adenoviral Shrna Vectors to Inhibit Tumor Cell Viability and Increase Chemotherapeutic Sensitivity to Gemcitabine in Bladder Cancer Cells
International Journal of Molecular Sciences Article Knockdown of RRM1 with Adenoviral shRNA Vectors to Inhibit Tumor Cell Viability and Increase Chemotherapeutic Sensitivity to Gemcitabine in Bladder Cancer Cells Xia Zhang 1, Rikiya Taoka 1,*, Dage Liu 2, Yuki Matsuoka 1, Yoichiro Tohi 1 , Yoshiyuki Kakehi 1 and Mikio Sugimoto 1 1 Department of Urology, Faculty of Medicine, Kagawa University, 1750-1 Ikenobe, Miki-cho, Kita-gun, Kagawa 761-0793, Japan; [email protected] (X.Z.); [email protected] (Y.M.); [email protected] (Y.T.); [email protected] (Y.K.); [email protected] (M.S.) 2 Department of General Thoracic Surgery, Faculty of Medicine, Kagawa University, 1750-1 Ikenobe, Miki-cho, Kita-gun, Kagawa 761-0793, Japan; [email protected] * Correspondence: [email protected]; Tel.: +81-87-891-2202 Abstract: RRM1—an important DNA replication/repair enzyme—is the primary molecular gem- citabine (GEM) target. High RRM1-expression associates with gemcitabine-resistance in various cancers and RRM1 inhibition may provide novel cancer treatment approaches. Our study eluci- dates how RRM1 inhibition affects cancer cell proliferation and influences gemcitabine-resistant bladder cancer cells. Of nine bladder cancer cell lines investigated, two RRM1 highly expressed cells, 253J and RT112, were selected for further experimentation. An RRM1-targeting shRNA was Citation: Zhang, X.; Taoka, R.; Liu, cloned into adenoviral vector, Ad-shRRM1. Gene and protein expression were investigated using D.; Matsuoka, Y.; Tohi, Y.; Kakehi, Y.; real-time PCR and western blotting. -

Supp Table 6.Pdf
Supplementary Table 6. Processes associated to the 2037 SCL candidate target genes ID Symbol Entrez Gene Name Process NM_178114 AMIGO2 adhesion molecule with Ig-like domain 2 adhesion NM_033474 ARVCF armadillo repeat gene deletes in velocardiofacial syndrome adhesion NM_027060 BTBD9 BTB (POZ) domain containing 9 adhesion NM_001039149 CD226 CD226 molecule adhesion NM_010581 CD47 CD47 molecule adhesion NM_023370 CDH23 cadherin-like 23 adhesion NM_207298 CERCAM cerebral endothelial cell adhesion molecule adhesion NM_021719 CLDN15 claudin 15 adhesion NM_009902 CLDN3 claudin 3 adhesion NM_008779 CNTN3 contactin 3 (plasmacytoma associated) adhesion NM_015734 COL5A1 collagen, type V, alpha 1 adhesion NM_007803 CTTN cortactin adhesion NM_009142 CX3CL1 chemokine (C-X3-C motif) ligand 1 adhesion NM_031174 DSCAM Down syndrome cell adhesion molecule adhesion NM_145158 EMILIN2 elastin microfibril interfacer 2 adhesion NM_001081286 FAT1 FAT tumor suppressor homolog 1 (Drosophila) adhesion NM_001080814 FAT3 FAT tumor suppressor homolog 3 (Drosophila) adhesion NM_153795 FERMT3 fermitin family homolog 3 (Drosophila) adhesion NM_010494 ICAM2 intercellular adhesion molecule 2 adhesion NM_023892 ICAM4 (includes EG:3386) intercellular adhesion molecule 4 (Landsteiner-Wiener blood group)adhesion NM_001001979 MEGF10 multiple EGF-like-domains 10 adhesion NM_172522 MEGF11 multiple EGF-like-domains 11 adhesion NM_010739 MUC13 mucin 13, cell surface associated adhesion NM_013610 NINJ1 ninjurin 1 adhesion NM_016718 NINJ2 ninjurin 2 adhesion NM_172932 NLGN3 neuroligin