Tropical Legume Trees and Their Soil-Mineral Microbiome: Biogeochemistry and Routes to Enhanced Mineral Access
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Patterns, Processes and Models of Microbial Recovery in a Chronosequence Following Reforestation of Reclaimed Mine Soils
PATTERNS, PROCESSES AND MODELS OF MICROBIAL RECOVERY IN A CHRONOSEQUENCE FOLLOWING REFORESTATION OF RECLAIMED MINE SOILS Shan Sun Dissertation submitted to the faculty of the Virginia Polytechnic Institute and State University in partial fulfillment of the requirements for the degree of Doctor of Philosophy In Crop and Soil Environmental Sciences Brian Badgley, Chair Song Li Brian Strahm Michael Strickland Carl Zipper Virginia Polytechnic Institute and State University Blacksburg, Virginia July 2017 Keywords: soil microorganism, recovery, chronosequence, reclaimed mine soils, amplicon sequencing, shotgun metagenomics, bacterial co-occurrence groups, artificial neural network PATTERNS, PROCESSES AND MODELS OF MICROBIAL RECOVERY IN A CHRONOSEQUENCE FOLLOWING REFORESTATION OF RECLAIMED MINE SOILS Shan Sun Abstract Soil microbial communities mediate important ecological processes and play essential roles in biogeochemical cycling. Ecosystem disturbances such as surface mining significantly alter soil microbial communities, which could lead to changes or impairment of ecosystem functions. Reforestation procedures were designed to accelerate the reestablishment of plant community and the recovery of the forest ecosystem after reclamation. However, the microbial recovery during reforestation has not been well studied even though this information is essential for evaluating ecosystem restoration success. In addition, the similar starting conditions of mining sites of different ages facilitate a chronosequence approach for studying decades-long microbial community change, which could help generalize theories about ecosystem succession. In this study, the recovery of microbial communities in a chronosequence of reclaimed mine sites spanning 30 years post reforestation along with unmined reference sites was analyzed using next-generation sequencing to characterize soil-microbial abundance, richness, taxonomic composition, interaction patterns and functional genes. -

Is Direct Seeding a Biologically Viable Strategy for Restoring Forest Ecosystems? Evidences from a Meta–Analysis
IS DIRECT SEEDING A BIOLOGICALLY VIABLE STRATEGY FOR RESTORING FOREST ECOSYSTEMS? EVIDENCES FROM A META–ANALYSIS 1* 2 2 Eliane Ceccon , Edgar J. González , Carlos Martorell 1 * Correspondence author: Centro Regional de Investigaciones Multidisciplinarias - Universidad Nacional Autónoma de México (UNAM), Av. Universidad s/n, Circuito 2, 62210, Col. Chamilpa, Cuernavaca, Morelos, Ciudad Universitaria de la UAEM, México. [email protected] 2 Departamento de Ecología y Recursos Naturales, Facultad de Ciencias, Universidad Nacional Autónoma de México, Av. Universidad 3000, Circuito Exterior S/N Delegación Coyoacán, C.P. 04510, Ciudad Universitaria, D.F. México. [email protected], [email protected] This article has been accepted for publication and undergone full peer review but has not been through the copyediting, typesetting, pagination and proofreading process which may lead to differences between this version and the Version of Record. Please cite this article as doi: 10.1002/ldr.2421 This article is protected by copyright. All rights reserved. Summary Ecological restoration has become an important technique for mitigating the human impacts on natural vegetation. Planting seedlings is the most common approach to regain lost forest cover. However, these activities require a large economic investment. Direct-seeding is considered a cheaper and easier alternative technique, in which tree seeds are introduced directly on the site rather than transplanting seedlings from nurseries. To evaluate the effectiveness of direct seeding, we conducted a comprehensive search of the literature using ―restoration‖, ―direct seeding‖ and ―sowing‖ as keywords, and we performed a meta-analysis using 30 papers and 89 species. We used two different measures of restoration success: seed germination probability and success probability (the chance that a seed germinates and survives until the end of the experiment). -

Exudate Flavonoids of Eight Species of Ceanothus (Rhamnaceae) Eckhard Wollenwebera,*, Marion Dörra, Bruce A
Exudate Flavonoids of Eight Species of Ceanothus (Rhamnaceae) Eckhard Wollenwebera,*, Marion Dörra, Bruce A. Bohmb, and James N. Roitmanc a Institut für Botanik der TU Darmstadt, Schnittspahnstrasse 3, D-64287 Darmstadt, Germany. Fax: 0049-6151/164630. E-mail: [email protected] b Botany Department, University of British Columbia, Vancouver, B.C., Canada c Western Regional Research Center, USDA-ARS, 800 Buchanan Street, Albany, CA 94710, U.S.A. * Author for correspondance and reprint requests Z. Naturforsch. 59c, 459Ð462 (2004); received April 26/May 13, 2004 Leaf glands of Ceanothus species excrete a lipophilic material that contains a variety of flavonoids. Most of these are aglycones, but some glycosides were also observed. Seven out of eight species exhibit flavonols, whereas flavones are excreted by only one species. Four species produce flavanones and dihydroflavonols; one excretes a remarkable quantity of fla- vonol glycosides. The exudate flavonoids thus form different patterns that might be charac- teristic for different Ceanothus species. Key words: Ceanothus, Rhamnaceae, Exudates, Flavonoids Introduction The genus Ceanothus (Rhamnaceae) contains about 60 species. These are deciduous or ever- green shrubs or rarely small trees that are some- Many species of the Rhamnaceae genus Ceano- times spiny. They occur chiefly in the Pacific coast thus bear multicellular glandular trichomes on region of North America from southwestern Can- their leaves, in particular along the margins. Fig. 1 ada to northern Mexico. Several species are culti- shows these glands on a leaf of C. papillosus and vated as ornamentals, and are available from local the typical shape of an individual C. hearstiorum nurseries. -

Brooklyn, Cloudland, Melsonby (Gaarraay)
BUSH BLITZ SPECIES DISCOVERY PROGRAM Brooklyn, Cloudland, Melsonby (Gaarraay) Nature Refuges Eubenangee Swamp, Hann Tableland, Melsonby (Gaarraay) National Parks Upper Bridge Creek Queensland 29 April–27 May · 26–27 July 2010 Australian Biological Resources Study What is Contents Bush Blitz? Bush Blitz is a four-year, What is Bush Blitz? 2 multi-million dollar Abbreviations 2 partnership between the Summary 3 Australian Government, Introduction 4 BHP Billiton and Earthwatch Reserves Overview 6 Australia to document plants Methods 11 and animals in selected properties across Australia’s Results 14 National Reserve System. Discussion 17 Appendix A: Species Lists 31 Fauna 32 This innovative partnership Vertebrates 32 harnesses the expertise of many Invertebrates 50 of Australia’s top scientists from Flora 62 museums, herbaria, universities, Appendix B: Threatened Species 107 and other institutions and Fauna 108 organisations across the country. Flora 111 Appendix C: Exotic and Pest Species 113 Fauna 114 Flora 115 Glossary 119 Abbreviations ANHAT Australian Natural Heritage Assessment Tool EPBC Act Environment Protection and Biodiversity Conservation Act 1999 (Commonwealth) NCA Nature Conservation Act 1992 (Queensland) NRS National Reserve System 2 Bush Blitz survey report Summary A Bush Blitz survey was conducted in the Cape Exotic vertebrate pests were not a focus York Peninsula, Einasleigh Uplands and Wet of this Bush Blitz, however the Cane Toad Tropics bioregions of Queensland during April, (Rhinella marina) was recorded in both Cloudland May and July 2010. Results include 1,186 species Nature Refuge and Hann Tableland National added to those known across the reserves. Of Park. Only one exotic invertebrate species was these, 36 are putative species new to science, recorded, the Spiked Awlsnail (Allopeas clavulinus) including 24 species of true bug, 9 species of in Cloudland Nature Refuge. -

Effects of Forest Fragmentation on Bottom-Up Control in Leaf-Cuttings Ants
Effects of forest fragmentation on bottom-up control in leaf-cuttings ants Dissertation zur Erlangung des naturwissenschaftlichen Doktorgrades Fachbereich Biologie Technische Universität Kaiserslautern vorgelegt von M.Sc. Pille Urbas Kaiserslautern, Dezember 2004 1. Gutachter: Prof. Dr. Burkhard Büdel 2. Gutachter: PD Dr. Jürgen Kusch Vorsitzender der Prüfungskommission: Prof. Dr. Matthias Hahn ACKNOWLEDGEMENTS I ACKNOWLEDGEMENTS I wish to thank my family for always being there; Joachim Gerhold who gave me great support and Jutta, Klaus and Markus Gerhold who decided to provide me with a second family; my supervisors Rainer Wirth, Burkhard Büdel and the department of Botany, University of Kaiserslautern for integrating me into the department and providing for such an interesting subject and the infrastructure to successfully work on it; the co-operators at the Federal University of Pernambuco (UFPE), Brazil - Inara Leal and Marcelo Tabarelli - for their assistance and interchange during my time overseas; the following students for the co-operatation in collecting and analysing data for some aspects of this study: Manoel Araújo (LAI and LCA leaf harvest), Ùrsula Costa (localization and size measurements of LCA colonies), Poliana Falcão (LCA diet breadth) and Nicole Meyer (tree density and DBH). Conservation International do Brasil, Centro de Estudos Ambientais do Nordeste and Usina Serra Grande for providing infrastructure during the field work; Marcia Nascimento, Lourinalda Silva and Lothar Bieber (UFPE) for sharing their laboratory, equipment and knowledge for chemical analyses; Jose Roberto Trigo (University of Campinas) for providing some special chemicals; my friends in Brazil Reisla Oliveira, Olivier Darrault, Cindy Garneau, Leonhard Krause, Edvaldo Florentino, Marcondes Oliveira and Alexandre Grillo for supporting me in a foreign land. -

Acacia Fimbriata Dwarf Crimson Blush 8 Eye on It During the Conference, Please Let Me Know
Australian Native Plants Society (Australia) Inc. ACACIA STUDY GROUP NEWSLETTER Group Leader and Newsletter Editor Seed Bank Curator Bill Aitchison Victoria Tanner 13 Conos Court, Donvale, Vic 3111 Phone (03) 98723583 Email: [email protected] No. 129 June 2015 ISSN 1035-4638 Contents Page From The Leader Dear Members From the Leader 1 It is now only a few months until the ANPSA Biennial Welcome 2 Conference being held in Canberra from 15-20 November. From Members and Readers 2 This is a great opportunity to catch up with some other Some Notes From Yallaroo 3 members of our Study Group, and of course to take part in Wattles With Minni Ritchi Bark 5 the great program put together by the organisers. Introduction of Australian Acacias Information relating to the Conference and details regarding to South America 6 registration are available on the Conference website Max’s Interesting Wattles 7 http://anpsa.org.au/conference2015. Our Study Group will An Acacia dealbata question from have a display at the Conference. If any Study Group Sweden 7 member who will be at the Conference could help with the Pre-treatment of Acacia Seeds 8 display, either in setting it up, or just in helping to keep an Acacia fimbriata dwarf Crimson Blush 8 eye on it during the Conference, please let me know. Books 9 Seed Bank 9 I am sure that many of our members will be aware of the Study Group Membership 10 Wattle Day Association, and the great work that it does in promoting National Wattle Day each year on 1 September. -

Successful Drug Discovery Informed by Actinobacterial Systematics
Successful Drug Discovery Informed by Actinobacterial Systematics Verrucosispora HPLC-DAD analysis of culture filtrate Structures of Abyssomicins Biological activity T DAD1, 7.382 (196 mAU,Up2) of 002-0101.D V. maris AB-18-032 mAU CH3 CH3 T extract H3C H3C Antibacterial activity (MIC): S. leeuwenhoekii C34 maris AB-18-032 175 mAU DAD1 A, Sig=210,10 150 C DAD1 B, Sig=230,10 O O DAD1 C, Sig=260,20 125 7 7 500 Rt 7.4 min DAD1 D, Sig=280,20 O O O O Growth inhibition of Gram-positive bacteria DAD1 , Sig=310,20 100 Abyssomicins DAD1 F, Sig=360,40 C 75 DAD1 G, Sig=435,40 Staphylococcus aureus (MRSA) 4 µg/ml DAD1 H, Sig=500,40 50 400 O O 25 O O Staphylococcus aureus (iVRSA) 13 µg/ml 0 CH CH3 300 400 500 nm 3 DAD1, 7.446 (300 mAU,Dn1) of 002-0101.D 300 mAU Mode of action: C HO atrop-C HO 250 atrop-C CH3 CH3 CH3 CH3 200 H C H C H C inhibitior of pABA biosynthesis 200 Rt 7.5 min H3C 3 3 3 Proximicin A Proximicin 150 HO O HO O O O O O O O O O A 100 O covalent binding to Cys263 of PabB 100 N 50 O O HO O O Sea of Japan B O O N O O (4-amino-4-deoxychorismate synthase) by 0 CH CH3 CH3 CH3 3 300 400 500 nm HO HO HO HO Michael addition -289 m 0 B D G H 2 4 6 8 10 12 14 16 min Newcastle Michael Goodfellow, School of Biology, University Newcastle University, Newcastle upon Tyne Atacama Desert In This Talk I will Consider: • Actinobacteria as a key group in the search for new therapeutic drugs. -

Native Plants Sixth Edition Sixth Edition AUSTRALIAN Native Plants Cultivation, Use in Landscaping and Propagation
AUSTRALIAN NATIVE PLANTS SIXTH EDITION SIXTH EDITION AUSTRALIAN NATIVE PLANTS Cultivation, Use in Landscaping and Propagation John W. Wrigley Murray Fagg Sixth Edition published in Australia in 2013 by ACKNOWLEDGEMENTS Reed New Holland an imprint of New Holland Publishers (Australia) Pty Ltd Sydney • Auckland • London • Cape Town Many people have helped us since 1977 when we began writing the first edition of Garfield House 86–88 Edgware Road London W2 2EA United Kingdom Australian Native Plants. Some of these folk have regrettably passed on, others have moved 1/66 Gibbes Street Chatswood NSW 2067 Australia to different areas. We endeavour here to acknowledge their assistance, without which the 218 Lake Road Northcote Auckland New Zealand Wembley Square First Floor Solan Road Gardens Cape Town 8001 South Africa various editions of this book would not have been as useful to so many gardeners and lovers of Australian plants. www.newhollandpublishers.com To the following people, our sincere thanks: Steve Adams, Ralph Bailey, Natalie Barnett, www.newholland.com.au Tony Bean, Lloyd Bird, John Birks, Mr and Mrs Blacklock, Don Blaxell, Jim Bourner, John Copyright © 2013 in text: John Wrigley Briggs, Colin Broadfoot, Dot Brown, the late George Brown, Ray Brown, Leslie Conway, Copyright © 2013 in map: Ian Faulkner Copyright © 2013 in photographs and illustrations: Murray Fagg Russell and Sharon Costin, Kirsten Cowley, Lyn Craven (Petraeomyrtus punicea photograph) Copyright © 2013 New Holland Publishers (Australia) Pty Ltd Richard Cummings, Bert -

Marine Science
Western Indian Ocean JOURNAL OF Marine Science Volume 18 | Issue 1 | Jan – Jun 2019 | ISSN: 0856-860X Chief Editor José Paula Western Indian Ocean JOURNAL OF Marine Science Chief Editor José Paula | Faculty of Sciences of University of Lisbon, Portugal Copy Editor Timothy Andrew Editorial Board Lena GIPPERTH Aviti MMOCHI Sweden Tanzania Serge ANDREFOUËT Johan GROENEVELD France Cosmas MUNGA South Africa Kenya Ranjeet BHAGOOLI Issufo HALO Mauritius South Africa/Mozambique Nyawira MUTHIGA Kenya Salomão BANDEIRA Christina HICKS Mozambique Australia/UK Brent NEWMAN Betsy Anne BEYMER-FARRIS Johnson KITHEKA South Africa USA/Norway Kenya Jan ROBINSON Jared BOSIRE Kassim KULINDWA Seycheles Kenya Tanzania Sérgio ROSENDO Atanásio BRITO Thierry LAVITRA Portugal Mozambique Madagascar Louis CELLIERS Blandina LUGENDO Melita SAMOILYS Kenya South Africa Tanzania Pascale CHABANET Joseph MAINA Max TROELL France Australia Sweden Published biannually Aims and scope: The Western Indian Ocean Journal of Marine Science provides an avenue for the wide dissem- ination of high quality research generated in the Western Indian Ocean (WIO) region, in particular on the sustainable use of coastal and marine resources. This is central to the goal of supporting and promoting sustainable coastal development in the region, as well as contributing to the global base of marine science. The journal publishes original research articles dealing with all aspects of marine science and coastal manage- ment. Topics include, but are not limited to: theoretical studies, oceanography, marine biology and ecology, fisheries, recovery and restoration processes, legal and institutional frameworks, and interactions/relationships between humans and the coastal and marine environment. In addition, Western Indian Ocean Journal of Marine Science features state-of-the-art review articles and short communications. -
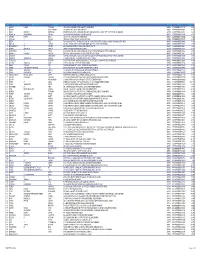
Section 124- Unpaid and Unclaimed Dividend
Sr No First Name Middle Name Last Name Address Pincode Folio Amount 1 ASHOK KUMAR GOLCHHA 305 ASHOKA CHAMBERS ADARSHNAGAR HYDERABAD 500063 0000000000B9A0011390 36.00 2 ADAMALI ABDULLABHOY 20, SUKEAS LANE, 3RD FLOOR, KOLKATA 700001 0000000000B9A0050954 150.00 3 AMAR MANOHAR MOTIWALA DR MOTIWALA'S CLINIC, SUNDARAM BUILDING VIKRAM SARABHAI MARG, OPP POLYTECHNIC AHMEDABAD 380015 0000000000B9A0102113 12.00 4 AMRATLAL BHAGWANDAS GANDHI 14 GULABPARK NEAR BASANT CINEMA CHEMBUR 400074 0000000000B9A0102806 30.00 5 ARVIND KUMAR DESAI H NO 2-1-563/2 NALLAKUNTA HYDERABAD 500044 0000000000B9A0106500 30.00 6 BIBISHAB S PATHAN 1005 DENA TOWER OPP ADUJAN PATIYA SURAT 395009 0000000000B9B0007570 144.00 7 BEENA DAVE 703 KRISHNA APT NEXT TO POISAR DEPOT OPP OUR LADY REMEDY SCHOOL S V ROAD, KANDIVILI (W) MUMBAI 400067 0000000000B9B0009430 30.00 8 BABULAL S LADHANI 9 ABDUL REHMAN STREET 3RD FLOOR ROOM NO 62 YUSUF BUILDING MUMBAI 400003 0000000000B9B0100587 30.00 9 BHAGWANDAS Z BAPHNA MAIN ROAD DAHANU DIST THANA W RLY MAHARASHTRA 401601 0000000000B9B0102431 48.00 10 BHARAT MOHANLAL VADALIA MAHADEVIA ROAD MANAVADAR GUJARAT 362630 0000000000B9B0103101 60.00 11 BHARATBHAI R PATEL 45 KRISHNA PARK SOC JASODA NAGAR RD NR GAUR NO KUVO PO GIDC VATVA AHMEDABAD 382445 0000000000B9B0103233 48.00 12 BHARATI PRAKASH HINDUJA 505 A NEEL KANTH 98 MARINE DRIVE P O BOX NO 2397 MUMBAI 400002 0000000000B9B0103411 60.00 13 BHASKAR SUBRAMANY FLAT NO 7 3RD FLOOR 41 SEA LAND CO OP HSG SOCIETY OPP HOTEL PRESIDENT CUFFE PARADE MUMBAI 400005 0000000000B9B0103985 96.00 14 BHASKER CHAMPAKLAL -

Chec List What Survived from the PLANAFLORO Project
Check List 10(1): 33–45, 2014 © 2014 Check List and Authors Chec List ISSN 1809-127X (available at www.checklist.org.br) Journal of species lists and distribution What survived from the PLANAFLORO Project: PECIES S Angiosperms of Rondônia State, Brazil OF 1* 2 ISTS L Samuel1 UniCarleialversity of Konstanz, and Narcísio Department C.of Biology, Bigio M842, PLZ 78457, Konstanz, Germany. [email protected] 2 Universidade Federal de Rondônia, Campus José Ribeiro Filho, BR 364, Km 9.5, CEP 76801-059. Porto Velho, RO, Brasil. * Corresponding author. E-mail: Abstract: The Rondônia Natural Resources Management Project (PLANAFLORO) was a strategic program developed in partnership between the Brazilian Government and The World Bank in 1992, with the purpose of stimulating the sustainable development and protection of the Amazon in the state of Rondônia. More than a decade after the PLANAFORO program concluded, the aim of the present work is to recover and share the information from the long-abandoned plant collections made during the project’s ecological-economic zoning phase. Most of the material analyzed was sterile, but the fertile voucher specimens recovered are listed here. The material examined represents 378 species in 234 genera and 76 families of angiosperms. Some 8 genera, 68 species, 3 subspecies and 1 variety are new records for Rondônia State. It is our intention that this information will stimulate future studies and contribute to a better understanding and more effective conservation of the plant diversity in the southwestern Amazon of Brazil. Introduction The PLANAFLORO Project funded botanical expeditions In early 1990, Brazilian Amazon was facing remarkably in different areas of the state to inventory arboreal plants high rates of forest conversion (Laurance et al. -

Rare Plant Survey of San Juan Public Lands, Colorado
Rare Plant Survey of San Juan Public Lands, Colorado 2005 Prepared by Colorado Natural Heritage Program 254 General Services Building Colorado State University Fort Collins CO 80523 Rare Plant Survey of San Juan Public Lands, Colorado 2005 Prepared by Peggy Lyon and Julia Hanson Colorado Natural Heritage Program 254 General Services Building Colorado State University Fort Collins CO 80523 December 2005 Cover: Imperiled (G1 and G2) plants of the San Juan Public Lands, top left to bottom right: Lesquerella pruinosa, Draba graminea, Cryptantha gypsophila, Machaeranthera coloradoensis, Astragalus naturitensis, Physaria pulvinata, Ipomopsis polyantha, Townsendia glabella, Townsendia rothrockii. Executive Summary This survey was a continuation of several years of rare plant survey on San Juan Public Lands. Funding for the project was provided by San Juan National Forest and the San Juan Resource Area of the Bureau of Land Management. Previous rare plant surveys on San Juan Public Lands by CNHP were conducted in conjunction with county wide surveys of La Plata, Archuleta, San Juan and San Miguel counties, with partial funding from Great Outdoors Colorado (GOCO); and in 2004, public lands only in Dolores and Montezuma counties, funded entirely by the San Juan Public Lands. Funding for 2005 was again provided by San Juan Public Lands. The primary emphases for field work in 2005 were: 1. revisit and update information on rare plant occurrences of agency sensitive species in the Colorado Natural Heritage Program (CNHP) database that were last observed prior to 2000, in order to have the most current information available for informing the revision of the Resource Management Plan for the San Juan Public Lands (BLM and San Juan National Forest); 2.