Intermediate Supercomputing Course Objectives
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Introduction to Multi-Threading and Vectorization Matti Kortelainen Larsoft Workshop 2019 25 June 2019 Outline
Introduction to multi-threading and vectorization Matti Kortelainen LArSoft Workshop 2019 25 June 2019 Outline Broad introductory overview: • Why multithread? • What is a thread? • Some threading models – std::thread – OpenMP (fork-join) – Intel Threading Building Blocks (TBB) (tasks) • Race condition, critical region, mutual exclusion, deadlock • Vectorization (SIMD) 2 6/25/19 Matti Kortelainen | Introduction to multi-threading and vectorization Motivations for multithreading Image courtesy of K. Rupp 3 6/25/19 Matti Kortelainen | Introduction to multi-threading and vectorization Motivations for multithreading • One process on a node: speedups from parallelizing parts of the programs – Any problem can get speedup if the threads can cooperate on • same core (sharing L1 cache) • L2 cache (may be shared among small number of cores) • Fully loaded node: save memory and other resources – Threads can share objects -> N threads can use significantly less memory than N processes • If smallest chunk of data is so big that only one fits in memory at a time, is there any other option? 4 6/25/19 Matti Kortelainen | Introduction to multi-threading and vectorization What is a (software) thread? (in POSIX/Linux) • “Smallest sequence of programmed instructions that can be managed independently by a scheduler” [Wikipedia] • A thread has its own – Program counter – Registers – Stack – Thread-local memory (better to avoid in general) • Threads of a process share everything else, e.g. – Program code, constants – Heap memory – Network connections – File handles -

Unit: 4 Processes and Threads in Distributed Systems
Unit: 4 Processes and Threads in Distributed Systems Thread A program has one or more locus of execution. Each execution is called a thread of execution. In traditional operating systems, each process has an address space and a single thread of execution. It is the smallest unit of processing that can be scheduled by an operating system. A thread is a single sequence stream within in a process. Because threads have some of the properties of processes, they are sometimes called lightweight processes. In a process, threads allow multiple executions of streams. Thread Structure Process is used to group resources together and threads are the entities scheduled for execution on the CPU. The thread has a program counter that keeps track of which instruction to execute next. It has registers, which holds its current working variables. It has a stack, which contains the execution history, with one frame for each procedure called but not yet returned from. Although a thread must execute in some process, the thread and its process are different concepts and can be treated separately. What threads add to the process model is to allow multiple executions to take place in the same process environment, to a large degree independent of one another. Having multiple threads running in parallel in one process is similar to having multiple processes running in parallel in one computer. Figure: (a) Three processes each with one thread. (b) One process with three threads. In former case, the threads share an address space, open files, and other resources. In the latter case, process share physical memory, disks, printers and other resources. -

Red Hat Developer Toolset 9 User Guide
Red Hat Developer Toolset 9 User Guide Installing and Using Red Hat Developer Toolset Last Updated: 2020-08-07 Red Hat Developer Toolset 9 User Guide Installing and Using Red Hat Developer Toolset Zuzana Zoubková Red Hat Customer Content Services Olga Tikhomirova Red Hat Customer Content Services [email protected] Supriya Takkhi Red Hat Customer Content Services Jaromír Hradílek Red Hat Customer Content Services Matt Newsome Red Hat Software Engineering Robert Krátký Red Hat Customer Content Services Vladimír Slávik Red Hat Customer Content Services Legal Notice Copyright © 2020 Red Hat, Inc. The text of and illustrations in this document are licensed by Red Hat under a Creative Commons Attribution–Share Alike 3.0 Unported license ("CC-BY-SA"). An explanation of CC-BY-SA is available at http://creativecommons.org/licenses/by-sa/3.0/ . In accordance with CC-BY-SA, if you distribute this document or an adaptation of it, you must provide the URL for the original version. Red Hat, as the licensor of this document, waives the right to enforce, and agrees not to assert, Section 4d of CC-BY-SA to the fullest extent permitted by applicable law. Red Hat, Red Hat Enterprise Linux, the Shadowman logo, the Red Hat logo, JBoss, OpenShift, Fedora, the Infinity logo, and RHCE are trademarks of Red Hat, Inc., registered in the United States and other countries. Linux ® is the registered trademark of Linus Torvalds in the United States and other countries. Java ® is a registered trademark of Oracle and/or its affiliates. XFS ® is a trademark of Silicon Graphics International Corp. -

Parallel Computing
Parallel Computing Announcements ● Midterm has been graded; will be distributed after class along with solutions. ● SCPD students: Midterms have been sent to the SCPD office and should be sent back to you soon. Announcements ● Assignment 6 due right now. ● Assignment 7 (Pathfinder) out, due next Tuesday at 11:30AM. ● Play around with graphs and graph algorithms! ● Learn how to interface with library code. ● No late submissions will be considered. This is as late as we're allowed to have the assignment due. Why Algorithms and Data Structures Matter Making Things Faster ● Choose better algorithms and data structures. ● Dropping from O(n2) to O(n log n) for large data sets will make your programs faster. ● Optimize your code. ● Try to reduce the constant factor in the big-O notation. ● Not recommended unless all else fails. ● Get a better computer. ● Having more memory and processing power can improve performance. ● New option: Use parallelism. How Your Programs Run Threads of Execution ● When running a program, that program gets a thread of execution (or thread). ● Each thread runs through code as normal. ● A program can have multiple threads running at the same time, each of which performs different tasks. ● A program that uses multiple threads is called multithreaded; writing a multithreaded program or algorithm is called multithreading. Threads in C++ ● The newest version of C++ (C++11) has libraries that support threading. ● To create a thread: ● Write the function that you want to execute. ● Construct an object of type thread to run that function. – Need header <thread> for this. ● That function will run in parallel alongside the original program. -

Threading SIMD and MIMD in the Multicore Context the Ultrasparc T2
Overview SIMD and MIMD in the Multicore Context Single Instruction Multiple Instruction ● (note: Tute 02 this Weds - handouts) ● Flynn’s Taxonomy Single Data SISD MISD ● multicore architecture concepts Multiple Data SIMD MIMD ● for SIMD, the control unit and processor state (registers) can be shared ■ hardware threading ■ SIMD vs MIMD in the multicore context ● however, SIMD is limited to data parallelism (through multiple ALUs) ■ ● T2: design features for multicore algorithms need a regular structure, e.g. dense linear algebra, graphics ■ SSE2, Altivec, Cell SPE (128-bit registers); e.g. 4×32-bit add ■ system on a chip Rx: x x x x ■ 3 2 1 0 execution: (in-order) pipeline, instruction latency + ■ thread scheduling Ry: y3 y2 y1 y0 ■ caches: associativity, coherence, prefetch = ■ memory system: crossbar, memory controller Rz: z3 z2 z1 z0 (zi = xi + yi) ■ intermission ■ design requires massive effort; requires support from a commodity environment ■ speculation; power savings ■ massive parallelism (e.g. nVidia GPGPU) but memory is still a bottleneck ■ OpenSPARC ● multicore (CMT) is MIMD; hardware threading can be regarded as MIMD ● T2 performance (why the T2 is designed as it is) ■ higher hardware costs also includes larger shared resources (caches, TLBs) ● the Rock processor (slides by Andrew Over; ref: Tremblay, IEEE Micro 2009 ) needed ⇒ less parallelism than for SIMD COMP8320 Lecture 2: Multicore Architecture and the T2 2011 ◭◭◭ • ◮◮◮ × 1 COMP8320 Lecture 2: Multicore Architecture and the T2 2011 ◭◭◭ • ◮◮◮ × 3 Hardware (Multi)threading The UltraSPARC T2: System on a Chip ● recall concurrent execution on a single CPU: switch between threads (or ● OpenSparc Slide Cast Ch 5: p79–81,89 processes) requires the saving (in memory) of thread state (register values) ● aggressively multicore: 8 cores, each with 8-way hardware threading (64 virtual ■ motivation: utilize CPU better when thread stalled for I/O (6300 Lect O1, p9–10) CPUs) ■ what are the costs? do the same for smaller stalls? (e.g. -

Download Chapter 3: "Configuring Your Project with Autoconf"
CONFIGURING YOUR PROJECT WITH AUTOCONF Come my friends, ’Tis not too late to seek a newer world. —Alfred, Lord Tennyson, “Ulysses” Because Automake and Libtool are essen- tially add-on components to the original Autoconf framework, it’s useful to spend some time focusing on using Autoconf without Automake and Libtool. This will provide a fair amount of insight into how Autoconf operates by exposing aspects of the tool that are often hidden by Automake. Before Automake came along, Autoconf was used alone. In fact, many legacy open source projects never made the transition from Autoconf to the full GNU Autotools suite. As a result, it’s not unusual to find a file called configure.in (the original Autoconf naming convention) as well as handwritten Makefile.in templates in older open source projects. In this chapter, I’ll show you how to add an Autoconf build system to an existing project. I’ll spend most of this chapter talking about the fundamental features of Autoconf, and in Chapter 4, I’ll go into much more detail about how some of the more complex Autoconf macros work and how to properly use them. Throughout this process, we’ll continue using the Jupiter project as our example. Autotools © 2010 by John Calcote Autoconf Configuration Scripts The input to the autoconf program is shell script sprinkled with macro calls. The input stream must also include the definitions of all referenced macros—both those that Autoconf provides and those that you write yourself. The macro language used in Autoconf is called M4. (The name means M, plus 4 more letters, or the word Macro.1) The m4 utility is a general-purpose macro language processor originally written by Brian Kernighan and Dennis Ritchie in 1977. -

Parallel Programming with Openmp
Parallel Programming with OpenMP OpenMP Parallel Programming Introduction: OpenMP Programming Model Thread-based parallelism utilized on shared-memory platforms Parallelization is either explicit, where programmer has full control over parallelization or through using compiler directives, existing in the source code. Thread is a process of a code is being executed. A thread of execution is the smallest unit of processing. Multiple threads can exist within the same process and share resources such as memory OpenMP Parallel Programming Introduction: OpenMP Programming Model Master thread is a single thread that runs sequentially; parallel execution occurs inside parallel regions and between two parallel regions, only the master thread executes the code. This is called the fork-join model: OpenMP Parallel Programming OpenMP Parallel Computing Hardware Shared memory allows immediate access to all data from all processors without explicit communication. Shared memory: multiple cpus are attached to the BUS all processors share the same primary memory the same memory address on different CPU's refer to the same memory location CPU-to-memory connection becomes a bottleneck: shared memory computers cannot scale very well OpenMP Parallel Programming OpenMP versus MPI OpenMP (Open Multi-Processing): easy to use; loop-level parallelism non-loop-level parallelism is more difficult limited to shared memory computers cannot handle very large problems An alternative is MPI (Message Passing Interface): require low-level programming; more difficult programming -

Intel's Hyper-Threading
Intel’s Hyper-Threading Cody Tinker & Christopher Valerino Introduction ● How to handle a thread, or the smallest portion of code that can be run independently, plays a core component in enhancing a program's parallelism. ● Due to the fact that modern computers process many tasks and programs simultaneously, techniques that allow for threads to be handled more efficiently to lower processor downtime are valuable. ● Motive is to reduce the number of idle resources of a processor. ● Examples of processors that use hyperthreading ○ Intel Xeon D-1529 ○ Intel i7-6785R ○ Intel Pentium D1517 Introduction ● Two main trends have been followed to increase parallelism: ○ Increase number of cores on a chip ○ Increase core throughput What is Multithreading? ● Executing more than one thread at a time ● Normally, an operating system handles a program by scheduling individual threads and then passing them to the processor. ● Two different types of hardware multithreading ○ Temporal ■ One thread per pipeline stage ○ Simultaneous (SMT) ■ Multiple threads per pipeline stage ● Intel’s hyper-threading is a SMT design Hyper-threading ● Hyper-threading is the hardware solution to increasing processor throughput by decreasing resource idle time. ● Allows multiple concurrent threads to be executed ○ Threads are interleaved so that resources not being used by one thread are used by others ○ Most processors are limited to 2 concurrent threads per physical core ○ Some do support 8 concurrent threads per physical core ● Needs the ability to fetch instructions from -
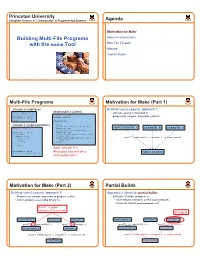
Building Multi-File Programs with the Make Tool
Princeton University Computer Science 217: Introduction to Programming Systems Agenda Motivation for Make Building Multi-File Programs Make Fundamentals with the make Tool Non-File Targets Macros Implicit Rules 1 2 Multi-File Programs Motivation for Make (Part 1) intmath.h (interface) Building testintmath, approach 1: testintmath.c (client) #ifndef INTMATH_INCLUDED • Use one gcc217 command to #define INTMATH_INCLUDED #include "intmath.h" int gcd(int i, int j); #include <stdio.h> preprocess, compile, assemble, and link int lcm(int i, int j); #endif int main(void) { int i; intmath.c (implementation) int j; printf("Enter the first integer:\n"); #include "intmath.h" scanf("%d", &i); testintmath.c intmath.h intmath.c printf("Enter the second integer:\n"); int gcd(int i, int j) scanf("%d", &j); { int temp; printf("Greatest common divisor: %d.\n", while (j != 0) gcd(i, j)); { temp = i % j; printf("Least common multiple: %d.\n", gcc217 testintmath.c intmath.c –o testintmath i = j; lcm(i, j); j = temp; return 0; } } return i; } Note: intmath.h is int lcm(int i, int j) #included into intmath.c { return (i / gcd(i, j)) * j; testintmath } and testintmath.c 3 4 Motivation for Make (Part 2) Partial Builds Building testintmath, approach 2: Approach 2 allows for partial builds • Preprocess, compile, assemble to produce .o files • Example: Change intmath.c • Link to produce executable binary file • Must rebuild intmath.o and testintmath • Need not rebuild testintmath.o!!! Recall: -c option tells gcc217 to omit link changed testintmath.c intmath.h intmath.c -

Lecture 3: a Simple Example, Tools, and CUDA Threads
ECE498AL Lecture 3: A Simple Example, Tools, and CUDA Threads © David Kirk/NVIDIA and Wen-mei W. Hwu, 2007-2009 1 ECE498AL, University of Illinois, Urbana-Champaign A Simple Running Example Matrix Multiplication • A simple matrix multiplication example that illustrates the basic features of memory and thread management in CUDA programs – Leave shared memory usage until later – Local, register usage – Thread ID usage – Memory data transfer API between host and device – Assume square matrix for simplicity © David Kirk/NVIDIA and Wen-mei W. Hwu, 2007-2009 2 ECE498AL, University of Illinois, Urbana-Champaign Programming Model: Square Matrix Multiplication Example • P = M * N of size WIDTH x WIDTH N • Without tiling: H T D – One thread calculates one element I W of P – M and N are loaded WIDTH times from global memory M P H T D I W WIDTH WIDTH © David Kirk/NVIDIA and Wen-mei W. Hwu, 2007-2009 3 ECE498AL, University of Illinois, Urbana-Champaign Memory Layout of a Matrix in C M0,0 M1,0 M2,0 M3,0 M0,1 M1,1 M2,1 M3,1 M0,2 M1,2 M2,2 M3,2 M0,3 M1,3 M2,3 M3,3 M M0,0 M1,0 M2,0 M3,0 M0,1 M1,1 M2,1 M3,1 M0,2 M1,2 M2,2 M3,2 M0,3 M1,3 M2,3 M3,3 © David Kirk/NVIDIA and Wen-mei W. Hwu, 2007-2009 4 ECE498AL, University of Illinois, Urbana-Champaign Step 1: Matrix Multiplication A Simple Host Version in C // Matrix multiplication on the (CPU) host in double precision void MatrixMulOnHost(float* M, float* N, float* P, int Width)N { k for (int i = 0; i < Width; ++i) H j T for (int j = 0; j < Width; ++j) { D I double sum = 0; W for (int k = 0; k < Width; ++k) { double a = M[i * width + k]; double b = N[k * width + j]; sum += a * b; M P } P[i * Width + j] = sum; i H } T D I } W k WIDTH WIDTH © David Kirk/NVIDIA and Wen-mei W. -

Thread Scheduling in Multi-Core Operating Systems Redha Gouicem
Thread Scheduling in Multi-core Operating Systems Redha Gouicem To cite this version: Redha Gouicem. Thread Scheduling in Multi-core Operating Systems. Computer Science [cs]. Sor- bonne Université, 2020. English. tel-02977242 HAL Id: tel-02977242 https://hal.archives-ouvertes.fr/tel-02977242 Submitted on 24 Oct 2020 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Ph.D thesis in Computer Science Thread Scheduling in Multi-core Operating Systems How to Understand, Improve and Fix your Scheduler Redha GOUICEM Sorbonne Université Laboratoire d’Informatique de Paris 6 Inria Whisper Team PH.D.DEFENSE: 23 October 2020, Paris, France JURYMEMBERS: Mr. Pascal Felber, Full Professor, Université de Neuchâtel Reviewer Mr. Vivien Quéma, Full Professor, Grenoble INP (ENSIMAG) Reviewer Mr. Rachid Guerraoui, Full Professor, École Polytechnique Fédérale de Lausanne Examiner Ms. Karine Heydemann, Associate Professor, Sorbonne Université Examiner Mr. Etienne Rivière, Full Professor, University of Louvain Examiner Mr. Gilles Muller, Senior Research Scientist, Inria Advisor Mr. Julien Sopena, Associate Professor, Sorbonne Université Advisor ABSTRACT In this thesis, we address the problem of schedulers for multi-core architectures from several perspectives: design (simplicity and correct- ness), performance improvement and the development of application- specific schedulers. -

Concurrent Computing
Concurrent Computing CSCI 201 Principles of Software Development Jeffrey Miller, Ph.D. [email protected] Outline • Threads • Multi-Threaded Code • CPU Scheduling • Program USC CSCI 201L Thread Overview ▪ Looking at the Windows taskbar, you will notice more than one process running concurrently ▪ Looking at the Windows task manager, you’ll see a number of additional processes running ▪ But how many processes are actually running at any one time? • Multiple processors, multiple cores, and hyper-threading will determine the actual number of processes running • Threads USC CSCI 201L 3/22 Redundant Hardware Multiple Processors Single-Core CPU ALU Cache Registers Multiple Cores ALU 1 Registers 1 Cache Hyper-threading has firmware logic that appears to the OS ALU 2 to contain more than one core when in fact only one physical core exists Registers 2 4/22 Programs vs Processes vs Threads ▪ Programs › An executable file residing in memory, typically in secondary storage ▪ Processes › An executing instance of a program › Sometimes this is referred to as a Task ▪ Threads › Any section of code within a process that can execute at what appears to be simultaneously › Shares the same resources allotted to the process by the OS kernel USC CSCI 201L 5/22 Concurrent Programming ▪ Multi-Threaded Programming › Multiple sections of a process or multiple processes executing at what appears to be simultaneously in a single core › The purpose of multi-threaded programming is to add functionality to your program ▪ Parallel Programming › Multiple sections of