Parallel Adaptations of Japanese Whiting, Sillago Japonica Under Temperature Stress
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Book of Abstracts
PICES-2016 25 Year of PICES: Celebrating the Past, Imagining the Future North Pacific Marine Science Organization November 2-13, 2016 San Diego, CA, USA Table of Contents Notes for Guidance � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � 5 Venue Floor Plan � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � 6 List of Sessions/Workshops � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � 9 Meeting Timetable � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � 10 PICES Structure � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � 12 PICES Acronyms � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � 13 Session/Workshop Schedules at a Glance � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � 15 List of Posters � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � 47 Sessions and Workshops Descriptions � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � � 63 Abstracts Oral Presentations (ordered by days) � � � � � � � � � � � � � � � � � � � � � -

Looking Beyond the Mortality of Bycatch: Sublethal Effects of Incidental Capture on Marine Animals
Biological Conservation 171 (2014) 61–72 Contents lists available at ScienceDirect Biological Conservation journal homepage: www.elsevier.com/locate/biocon Review Looking beyond the mortality of bycatch: sublethal effects of incidental capture on marine animals a, a a,b b a Samantha M. Wilson ⇑, Graham D. Raby , Nicholas J. Burnett , Scott G. Hinch , Steven J. Cooke a Fish Ecology and Conservation Physiology Laboratory, Department of Biology and Institute of Environmental Sciences, Carleton University, Ottawa, ON, Canada b Pacific Salmon Ecology and Conservation Laboratory, Department of Forest and Conservation Sciences, University of British Columbia, Vancouver, BC, Canada article info abstract Article history: There is a widely recognized need to understand and reduce the incidental effects of marine fishing on Received 14 August 2013 non-target animals. Previous research on marine bycatch has largely focused on simply quantifying mor- Received in revised form 10 January 2014 tality. However, much less is known about the organism-level sublethal effects, including the potential Accepted 13 January 2014 for behavioural alterations, physiological and energetic costs, and associated reductions in feeding, growth, or reproduction (i.e., fitness) which can occur undetected following escape or release from fishing gear. We reviewed the literature and found 133 marine bycatch papers that included sublethal endpoints Keywords: such as physiological disturbance, behavioural impairment, injury, reflex impairment, and effects on RAMP reproduction, -

ORGANIC CHEMICAL TOXICOLOGY of FISHES This Is Volume 33 in The
ORGANIC CHEMICAL TOXICOLOGY OF FISHES This is Volume 33 in the FISH PHYSIOLOGY series Edited by Anthony P. Farrell and Colin J. Brauner Honorary Editors: William S. Hoar and David J. Randall A complete list of books in this series appears at the end of the volume ORGANIC CHEMICAL TOXICOLOGY OF FISHES Edited by KEITH B. TIERNEY Department of Biological Sciences University of Alberta Edmonton, Alberta Canada ANTHONY P. FARRELL Department of Zoology, and Faculty of Land and Food Systems The University of British Columbia Vancouver, British Columbia Canada COLIN J. BRAUNER Department of Zoology The University of British Columbia Vancouver, British Columbia Canada AMSTERDAM BOSTON HEIDELBERG LONDON NEW YORK OXFORD PARIS SAN DIEGO SAN FRANCISCO SINGAPORE SYDNEY TOKYO Academic Press is an imprint of Elsevier Academic Press is an imprint of Elsevier 32 Jamestown Road, London NW1 7BY, UK 225 Wyman Street, Waltham, MA 02451, USA 525 B Street, Suite 1800, San Diego, CA 92101-4495, USA Copyright r 2014 Elsevier Inc. All rights reserved The cover illustrates the diversity of effects an example synthetic organic water pollutant can have on fish. The chemical shown is 2,4-D, an herbicide that can be found in streams near urbanization and agriculture. The fish shown is one that can live in such streams: rainbow trout (Oncorhynchus mykiss). The effect shown on the left is the ability of 2,4-D (yellow line) to stimulate olfactory sensory neurons vs. control (black line) (measured as an electro- olfactogram; EOG). The effect shown on the right is the ability of 2,4-D to induce the expression of an egg yolk precursor protein (vitellogenin) in male fish. -
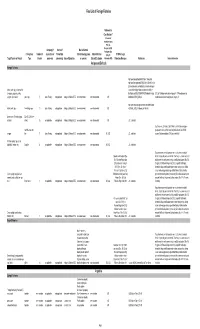
2018 Final LOFF W/ Ref and Detailed Info
Final List of Foreign Fisheries Rationale for Classification ** (Presence of mortality or injury (P/A), Co- Occurrence (C/O), Company (if Source of Marine Mammal Analogous Gear Fishery/Gear Number of aquaculture or Product (for Interactions (by group Marine Mammal (A/G), No RFMO or Legal Target Species or Product Type Vessels processor) processing) Area of Operation or species) Bycatch Estimates Information (N/I)) Protection Measures References Detailed Information Antigua and Barbuda Exempt Fisheries http://www.fao.org/fi/oldsite/FCP/en/ATG/body.htm http://www.fao.org/docrep/006/y5402e/y5402e06.htm,ht tp://www.tradeboss.com/default.cgi/action/viewcompan lobster, rock, spiny, demersal fish ies/searchterm/spiny+lobster/searchtermcondition/1/ , (snappers, groupers, grunts, ftp://ftp.fao.org/fi/DOCUMENT/IPOAS/national/Antigua U.S. LoF Caribbean spiny lobster trap/ pot >197 None documented, surgeonfish), flounder pots, traps 74 Lewis Fishing not applicable Antigua & Barbuda EEZ none documented none documented A/G AndBarbuda/NPOA_IUU.pdf Caribbean mixed species trap/pot are category III http://www.nmfs.noaa.gov/pr/interactions/fisheries/tabl lobster, rock, spiny free diving, loops 19 Lewis Fishing not applicable Antigua & Barbuda EEZ none documented none documented A/G e2/Atlantic_GOM_Caribbean_shellfish.html Queen conch (Strombus gigas), Dive (SCUBA & free molluscs diving) 25 not applicable not applicable Antigua & Barbuda EEZ none documented none documented A/G U.S. trade data Southeastern U.S. Atlantic, Gulf of Mexico, and Caribbean snapper- handline, hook and grouper and other reef fish bottom longline/hook-and-line/ >5,000 snapper line 71 Lewis Fishing not applicable Antigua & Barbuda EEZ none documented none documented N/I, A/G U.S. -
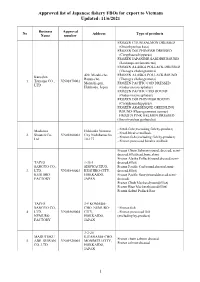
Approved List of Japanese Fishery Fbos for Export to Vietnam Updated: 11/6/2021
Approved list of Japanese fishery FBOs for export to Vietnam Updated: 11/6/2021 Business Approval No Address Type of products Name number FROZEN CHUM SALMON DRESSED (Oncorhynchus keta) FROZEN DOLPHINFISH DRESSED (Coryphaena hippurus) FROZEN JAPANESE SARDINE ROUND (Sardinops melanostictus) FROZEN ALASKA POLLACK DRESSED (Theragra chalcogramma) 420, Misaki-cho, FROZEN ALASKA POLLACK ROUND Kaneshin Rausu-cho, (Theragra chalcogramma) 1. Tsuyama CO., VN01870001 Menashi-gun, FROZEN PACIFIC COD DRESSED LTD Hokkaido, Japan (Gadus macrocephalus) FROZEN PACIFIC COD ROUND (Gadus macrocephalus) FROZEN DOLPHIN FISH ROUND (Coryphaena hippurus) FROZEN ARABESQUE GREENLING ROUND (Pleurogrammus azonus) FROZEN PINK SALMON DRESSED (Oncorhynchus gorbuscha) - Fresh fish (excluding fish by-product) Maekawa Hokkaido Nemuro - Fresh bivalve mollusk. 2. Shouten Co., VN01860002 City Nishihamacho - Frozen fish (excluding fish by-product) Ltd 10-177 - Frozen processed bivalve mollusk Frozen Chum Salmon (round, dressed, semi- dressed,fillet,head,bone,skin) Frozen Alaska Pollack(round,dressed,semi- TAIYO 1-35-1 dressed,fillet) SANGYO CO., SHOWACHUO, Frozen Pacific Cod(round,dressed,semi- 3. LTD. VN01840003 KUSHIRO-CITY, dressed,fillet) KUSHIRO HOKKAIDO, Frozen Pacific Saury(round,dressed,semi- FACTORY JAPAN dressed) Frozen Chub Mackerel(round,fillet) Frozen Blue Mackerel(round,fillet) Frozen Salted Pollack Roe TAIYO 3-9 KOMABA- SANGYO CO., CHO, NEMURO- - Frozen fish 4. LTD. VN01860004 CITY, - Frozen processed fish NEMURO HOKKAIDO, (excluding by-product) FACTORY JAPAN -

Chinese Red Swimming Crab (Portunus Haanii) Fishery Improvement Project (FIP) in Dongshan, China (August-December 2018)
Chinese Red Swimming Crab (Portunus haanii) Fishery Improvement Project (FIP) in Dongshan, China (August-December 2018) Prepared by Min Liu & Bai-an Lin Fish Biology Laboratory College of Ocean and Earth Sciences, Xiamen University March 2019 Contents 1. Introduction........................................................................................................ 5 2. Materials and Methods ...................................................................................... 6 2.1. Study site and survey frequency .................................................................... 6 2.2. Sample collection .......................................................................................... 7 2.3. Species identification................................................................................... 10 2.4. Sample measurement ................................................................................... 11 2.5. Interviews.................................................................................................... 13 2.6. Estimation of annual capture volume of Portunus haanii ............................. 15 3. Results .............................................................................................................. 15 3.1. Species diversity.......................................................................................... 15 3.1.1. Species composition .............................................................................. 15 3.1.2. ETP species ......................................................................................... -

Acta Botanica Brasilica - 35(1): 22-36
Acta Botanica Brasilica - 35(1): 22-36. January-March 2021. doi: 10.1590/0102-33062020abb0188 Predicting the potential distribution of aquatic herbaceous plants in oligotrophic Central Amazonian wetland ecosystems Aline Lopes1, 2* , Layon Oreste Demarchi2, 3 , Augusto Cesar Franco1 , Aurélia Bentes Ferreira2, 3 , Cristiane Silva Ferreira1 , Florian Wittmann2, 4 , Ivone Neri Santiago2 , Jefferson da Cruz2, 5 , Jeisiane Santos da Silva2, 3 , Jochen Schöngart2, 3 , Sthefanie do Nascimento Gomes de Souza2 and Maria Teresa Fernandez Piedade2, 3 Received: May 25, 2020 Accepted: November 13, 2020 ABSTRACT . Aquatic herbaceous plants are especially suitable for mapping environmental variability in wetlands, as they respond quickly to environmental gradients and are good indicators of habitat preference. We describe the composition of herbaceous species in two oligotrophic wetland ecosystems, floodplains along black-water rivers (igapó) and wetlands upon hydromorphic sand soils (campinarana) in the Parque Nacional do Jaú and the Reserva de Desenvolvimento Sustentável Uatumã in Central Amazonia, both protected areas. We tested for the potential distribution range (PDR) of the most frequent species of these ecosystems, which are the ones that occurred in at least two of the sampled wetlands, using species distribution models (SDMs). In total, 98 aquatic herbaceous species were recorded, of which 63 occurred in igapós and 44 in campinaranas. Most igapó species had ample PDRs across the Neotropics, while most campinaranas species were restricted to the Amazon Basin. These results are congruent with studies that described similar distribution patterns for tree and bird species, which emphasizes a high degree of endemism in Amazonian campinarana. However, we also found differences in the potential distribution of species between the two protected areas, indicating high environmental variability of oligotrophic ecosystems that deserve further investigation to develop effective measures for their conservation and protection. -

Title Importance of Estuaries and Rivers for the Coastal Fish, Temperate
Importance of estuaries and rivers for the coastal fish, Title temperate seabass Lateolabrax japonicus( Dissertation_全文 ) Author(s) Fuji, Taiki Citation 京都大学 Issue Date 2014-03-24 URL https://doi.org/10.14989/doctor.k18337 Right 許諾条件により本文は2015-03-01に公開 Type Thesis or Dissertation Textversion ETD Kyoto University Importance of estuaries and rivers for the coastal fish, temperate seabass Lateolabrax japonicus 2014 Taiki Fuji Contents Abstract...............................................................................................................1 Chapter 1: General introduction……………………….…………………………6 Chapter 2: Impacts of river discharge on larval temperate seabass recruitment towards coastal nursery grounds in the Tango Sea……………13 Chapter 3: Distributions and size ranges of juvenile temperate seabass in the Tango Sea……………………………………………………………………………24 Chapter 4: River ascending mechanisms of juvenile temperate seabass in the stratified Yura River estuary……....................................................................42 Chapter 5: Feeding ecology of juvenile temperate seabass in the Yura River estuary……………………………………………………………………………...64 Chapter 6: Migration ecology of juvenile temperate seabass: a carbon stable isotope approach…………………………………….……………………….………94 Chapter 7: Growth and migration patterns of juvenile temperate seabass in the Yura River estuary - combination of stable isotope ratio and otolith microstructure analyses………………………………………………………...109 Chapter 8: Main factor of the mortality during the estuarine juvenile stage of temperate -

Selenium Content in Seafood in Japan
Nutrients 2013, 5, 388-395; doi:10.3390/nu5020388 OPEN ACCESS nutrients ISSN 2072-6643 www.mdpi.com/journal/nutrients Article Selenium Content in Seafood in Japan Yumiko Yamashita *, Michiaki Yamashita and Haruka Iida National Research Institute of Fisheries Science, Kanazawa, Yokohama, Kanagawa, 236-8648, Japan; E-Mails: [email protected] (M.Y.); [email protected] (H.I.) * Author to whom correspondence should be addressed; E-Mail: [email protected]; Tel.: +81-45-788-7665; Fax: +81-45-788-5001. Received: 5 January 2013; in revised form: 24 January 2013 / Accepted: 25 January 2013 / Published: 31 January 2013 Abstract: Selenium is an essential micronutrient for humans, and seafood is one of the major selenium sources, as well as red meat, grains, eggs, chicken, liver and garlic. A substantial proportion of the total amount of selenium is present as selenium containing imidazole compound, selenoneine, in the muscles of ocean fish. In order to characterize the selenium content in seafood, the total selenium levels were measured in the edible portions of commercially important fish and shellfish species. Among the tested edible portions, alfonsino muscle had the highest selenium levels (concentration of 1.27 mg/kg tissue). High levels of selenium (1.20–1.07 mg/kg) were also found in the salted ovary products of mullet and Pacific herring. In other fish muscles, the selenium levels ranged between 0.12 and 0.77 mg/kg tissue. The selenium levels were closely correlated with the mercury levels in the white and red muscles in alfonsino. The selenium content in spleen, blood, hepatopancreas, heart, red muscle, white muscle, brain, ovary and testis ranged between 1.10 and 24.8 mg/kg tissue in alfonsino. -

Larval Development of King George
Abstract.---The larval develop ment of Sillaginodes punctata, Larval development of King George Sillago bassensis, and Sillago schomburgkii is described based on whiting, Sillaginodes punctata, both field-collected and laboratory reared material. Larvae of the school whiting, Sillago bassensis, three species can be separated based on a combination ofpigment and yellow fin whiting, and meristic characters, including extent and appearance of dorsal Sillago schomburgkii midline pigment, lateral pigment on the tail, presence or absence of pigment above the notochord tip, (Percoidei: Sillaginidae), from myomere number, extent and tim ing of gut coiling, and size at flex South Australian waters ion. The most useful meristic char acter across the range ofspecimens Barry D. Bruce was number of myomeres. Sillagi nodes punctata with 42-45 myo South Australian Department of Fisheries meres are easily distinguished GPO Box J625. Adelaide. South Australia 500 J from Sillago schomburgkii with Present address: (SIRO Division of Fisheries. 36-38, and from S. bassensis with GPO Box J538. Hobart. Tasmania. Australia 700 J. 32-35. The timing of gut coiling and its subsequent effect on anus position differed both among the three species examined here and from that previously reported for sillaginid larvae in general. Timing The perciform family Sillaginidae sidered subspecies of S. bassensis of gut coiling and extent of anus (whiting and sand smelts) consists (McKay, 19921. All four species are migration are not useful characters of three genera, three subgenera, widely distributed in southernAus for the identification of temperate and thirty-one species of small to tralia and form the basis for impor Australian sillaginids at the fam moderately sized fishes found pri tant commercial fisheries across ily level but are useful on a specific level. -

(Danio Rerio) Embryos/Larvae at Environmentally Relevant Levels
Chemosphere 189 (2017) 498e506 Contents lists available at ScienceDirect Chemosphere journal homepage: www.elsevier.com/locate/chemosphere Tributyltin induces premature hatching and reduces locomotor activity in zebrafish (Danio rerio) embryos/larvae at environmentally relevant levels * Xuefang Liang a, b, Christopher L. Souders II b, Jiliang Zhang c, Christopher J. Martyniuk b, a School of Ecology and Environment, Inner Mongolia University, Hohhot, 010021, China b Center for Environmental and Human Toxicology, Department of Physiological Sciences, College of Veterinary Medicine, UF Genetics Institute, Interdisciplinary Program in Biomedical Sciences Neuroscience, University of Florida, Gainesville, FL, 32611, USA c Henan Open Laboratory of Key Subjects of Environmental and Animal Products Safety, College of Animal Science and Technology, Henan University of Science and Technology, Henan, China highlights Exposure to 1 nM TBT induced premature hatching in zebrafish. TBT at 1 nM inhibited locomotor behaviors of zebrafish larvae. Mitochondrial bioenergetics were not affected by TBT. Genes related to muscle function and dopamine signaling were not altered by TBT. article info abstract Article history: Tributyltin (TBT) is an organotin compound that is the active ingredient of many biocides and antifouling Received 27 July 2017 agents. In addition to its well established role as an endocrine disruptor, TBT is also associated with Received in revised form adverse effects on the nervous system and behavior. In this study, zebrafish (Danio rerio) embryos were 18 September 2017 exposed to environmentally relevant concentrations of TBT (0.01, 0.1, 1 nM) to determine how low levels Accepted 19 September 2017 affected development and behavior. Fish exposed to 1 nM TBT hatched earlier when compared to con- Available online 20 September 2017 trols. -

Modelling the Effects of Climate Change on the Distribution of Endangered Cypripedium Japonicum in China
Article Modelling the Effects of Climate Change on the Distribution of Endangered Cypripedium japonicum in China Yadong Xu 1,2, Yi Huang 1,2 , Huiru Zhao 1,2, Meiling Yang 1,2, Yuqi Zhuang 1,2 and Xinping Ye 1,2,* 1 College of Life Sciences, Shaanxi Normal University, Xi’an 710119, China; [email protected] (Y.X.); [email protected] (Y.H.); [email protected] (H.Z.); [email protected] (M.Y.); [email protected] (Y.Z.) 2 Research Center for UAV Remote Sensing, Shaanxi Normal University, Xi’an 710119, China * Correspondence: [email protected]; Tel.: +86-029-8531-0266 Abstract: Cypripedium japonicum is an endangered terrestrial orchid species with high ornamental and medicinal value. As global warming continues to intensify, the survival of C. japonicum will be further challenged. Understanding the impact of climate change on its potential distribution is of great significance to conserve this species. In this study, we established an ensemble species distribution model based on occurrence records of C. japonicum and 13 environmental variables to predict its potential distribution under current and future climatic conditions. The results show that the true skill statistic (TSS), Cohen’s kappa statistic (Kappa), and the area under the receiver operating characteristic curve (AUC) values of the ensemble model were 0.968, 0.906, and 0.995, respectively, providing more robust predictions. The key environmental variables affecting the distribution of C. japonicum were the precipitation in the warmest quarter (Bio18) and the mean temperature in the driest quarter (Bio9). Under future climatic conditions, the total suitable habitat of C.