Growth Kinetics and Constraints Related to Metabolic Diversity And
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Washington Division of Geology and Earth Resources Open File Report
l 122 EARTHQUAKES AND SEISMOLOGY - LEGAL ASPECTS OPEN FILE REPORT 92-2 EARTHQUAKES AND Ludwin, R. S.; Malone, S. D.; Crosson, R. EARTHQUAKES AND SEISMOLOGY - LEGAL S.; Qamar, A. I., 1991, Washington SEISMOLOGY - 1946 EVENT ASPECTS eanhquak:es, 1985. Clague, J. J., 1989, Research on eanh- Ludwin, R. S.; Qamar, A. I., 1991, Reeval Perkins, J. B.; Moy, Kenneth, 1989, Llabil quak:e-induced ground failures in south uation of the 19th century Washington ity of local government for earthquake western British Columbia [abstract). and Oregon eanhquake catalog using hazards and losses-A guide to the law Evans, S. G., 1989, The 1946 Mount Colo original accounts-The moderate sized and its impacts in the States of Califor nel Foster rock avalanches and auoci earthquake of May l, 1882 [abstract). nia, Alaska, Utah, and Washington; ated displacement wave, Vancouver Is Final repon. Maley, Richard, 1986, Strong motion accel land, British Columbia. erograph stations in Oregon and Wash Hasegawa, H. S.; Rogers, G. C., 1978, EARTHQUAKES AND ington (April 1986). Appendix C Quantification of the magnitude 7.3, SEISMOLOGY - NETWORKS Malone, S. D., 1991, The HAWK seismic British Columbia earthquake of June 23, AND CATALOGS data acquisition and analysis system 1946. [abstract). Berg, J. W., Jr.; Baker, C. D., 1963, Oregon Hodgson, E. A., 1946, British Columbia eanhquak:es, 1841 through 1958 [ab Milne, W. G., 1953, Seismological investi earthquake, June 23, 1946. gations in British Columbia (abstract). stract). Hodgson, J. H.; Milne, W. G., 1951, Direc Chan, W.W., 1988, Network and array anal Munro, P. S.; Halliday, R. J.; Shannon, W. -

The 2014 Golden Gate National Parks Bioblitz - Data Management and the Event Species List Achieving a Quality Dataset from a Large Scale Event
National Park Service U.S. Department of the Interior Natural Resource Stewardship and Science The 2014 Golden Gate National Parks BioBlitz - Data Management and the Event Species List Achieving a Quality Dataset from a Large Scale Event Natural Resource Report NPS/GOGA/NRR—2016/1147 ON THIS PAGE Photograph of BioBlitz participants conducting data entry into iNaturalist. Photograph courtesy of the National Park Service. ON THE COVER Photograph of BioBlitz participants collecting aquatic species data in the Presidio of San Francisco. Photograph courtesy of National Park Service. The 2014 Golden Gate National Parks BioBlitz - Data Management and the Event Species List Achieving a Quality Dataset from a Large Scale Event Natural Resource Report NPS/GOGA/NRR—2016/1147 Elizabeth Edson1, Michelle O’Herron1, Alison Forrestel2, Daniel George3 1Golden Gate Parks Conservancy Building 201 Fort Mason San Francisco, CA 94129 2National Park Service. Golden Gate National Recreation Area Fort Cronkhite, Bldg. 1061 Sausalito, CA 94965 3National Park Service. San Francisco Bay Area Network Inventory & Monitoring Program Manager Fort Cronkhite, Bldg. 1063 Sausalito, CA 94965 March 2016 U.S. Department of the Interior National Park Service Natural Resource Stewardship and Science Fort Collins, Colorado The National Park Service, Natural Resource Stewardship and Science office in Fort Collins, Colorado, publishes a range of reports that address natural resource topics. These reports are of interest and applicability to a broad audience in the National Park Service and others in natural resource management, including scientists, conservation and environmental constituencies, and the public. The Natural Resource Report Series is used to disseminate comprehensive information and analysis about natural resources and related topics concerning lands managed by the National Park Service. -

Diversity of Understudied Archaeal and Bacterial Populations of Yellowstone National Park: from Genes to Genomes Daniel Colman
University of New Mexico UNM Digital Repository Biology ETDs Electronic Theses and Dissertations 7-1-2015 Diversity of understudied archaeal and bacterial populations of Yellowstone National Park: from genes to genomes Daniel Colman Follow this and additional works at: https://digitalrepository.unm.edu/biol_etds Recommended Citation Colman, Daniel. "Diversity of understudied archaeal and bacterial populations of Yellowstone National Park: from genes to genomes." (2015). https://digitalrepository.unm.edu/biol_etds/18 This Dissertation is brought to you for free and open access by the Electronic Theses and Dissertations at UNM Digital Repository. It has been accepted for inclusion in Biology ETDs by an authorized administrator of UNM Digital Repository. For more information, please contact [email protected]. Daniel Robert Colman Candidate Biology Department This dissertation is approved, and it is acceptable in quality and form for publication: Approved by the Dissertation Committee: Cristina Takacs-Vesbach , Chairperson Robert Sinsabaugh Laura Crossey Diana Northup i Diversity of understudied archaeal and bacterial populations from Yellowstone National Park: from genes to genomes by Daniel Robert Colman B.S. Biology, University of New Mexico, 2009 DISSERTATION Submitted in Partial Fulfillment of the Requirements for the Degree of Doctor of Philosophy Biology The University of New Mexico Albuquerque, New Mexico July 2015 ii DEDICATION I would like to dedicate this dissertation to my late grandfather, Kenneth Leo Colman, associate professor of Animal Science in the Wool laboratory at Montana State University, who even very near the end of his earthly tenure, thought it pertinent to quiz my knowledge of oxidized nitrogen compounds. He was a man of great curiosity about the natural world, and to whom I owe an acknowledgement for his legacy of intellectual (and actual) wanderlust. -

Genomics and Energy and Environmental Science Poster 2011
Omics: Exploring the Molecular Universe within Biological Systems Instead of studying one or a few genes or proteins at a time, “omics” collectively Proteomics is the analysis of the proteome—the complete set describes the comprehensive analysis of genes, RNA transcripts, proteins, of proteins expressed by a cell or population of cells. Proteins, metabolites, and other molecules present in a biological system. Ongoing the workhorse molecules of life, catalyze biochemical reactions; advances in computing power and automated technologies for DNA sequencing provide structural support; and recognize, bind, or transport and experiments continue to improve our ability to analyze increasing numbers other molecules throughout the cell. Hundreds of dierent types of molecules and how they function as a system. Systems biology integrates the of proteins can be expressed at a time, and most are part of large data from various omic analyses using computational tools to build predictive complexes made up of models of biological systems. many proteins and other molecules. Transcriptomics is the analysis of the transcriptome—the complete set of RNA Hundreds to molecules present in a cell or population of thousands of different cells. RNA, which is much less stable than protein types exist within DNA, is constantly being synthesized and a cell. This large subunit of a ribosome contains about then broken down to facilitate rapid 3,000 RNA nucleotides changes in paerns of protein expression (gray) and 30 protein that occur as an organism dynamically chains (gold). responds to its environment. In addition to the three major classes of RNA (mRNA, tRNA, and ribosomal RNA), single- stranded RNA is very exible and can fold into complex shapes that carry out specic functions. -

Biological Diversity in the Patent System
Biological Diversity in the Patent System Paul Oldham1,2*, Stephen Hall1,3, Oscar Forero1,4 1 ESRC Centre for Economic and Social Aspects of Genomics (Cesagen), Lancaster University, Lancaster, United Kingdom, 2 Institute of Advanced Studies, United Nations University, Yokohama, Japan, 3 One World Analytics, Lancaster, United Kingdom, 4 Centre for Development, Environment and Policy, SOAS, University of London, London, United Kingdom Abstract Biological diversity in the patent system is an enduring focus of controversy but empirical analysis of the presence of biodiversity in the patent system has been limited. To address this problem we text mined 11 million patent documents for 6 million Latin species names from the Global Names Index (GNI) established by the Global Biodiversity Information Facility (GBIF) and Encyclopedia of Life (EOL). We identified 76,274 full Latin species names from 23,882 genera in 767,955 patent documents. 25,595 species appeared in the claims section of 136,880 patent documents. This reveals that human innovative activity involving biodiversity in the patent system focuses on approximately 4% of taxonomically described species and between 0.8–1% of predicted global species. In this article we identify the major features of the patent landscape for biological diversity by focusing on key areas including pharmaceuticals, neglected diseases, traditional medicines, genetic engineering, foods, biocides, marine genetic resources and Antarctica. We conclude that the narrow focus of human innovative activity and ownership of genetic resources is unlikely to be in the long term interest of humanity. We argue that a broader spectrum of biodiversity needs to be opened up to research and development based on the principles of equitable benefit-sharing, respect for the objectives of the Convention on Biological Diversity, human rights and ethics. -

Phylogenetics of Archaeal Lipids Amy Kelly 9/27/2006 Outline
Phylogenetics of Archaeal Lipids Amy Kelly 9/27/2006 Outline • Phlogenetics of Archaea • Phlogenetics of archaeal lipids • Papers Phyla • Two? main phyla – Euryarchaeota • Methanogens • Extreme halophiles • Extreme thermophiles • Sulfate-reducing – Crenarchaeota • Extreme thermophiles – Korarchaeota? • Hyperthermophiles • indicated only by environmental DNA sequences – Nanoarchaeum? • N. equitans a fast evolving euryarchaeal lineage, not novel, early diverging archaeal phylum – Ancient archael group? • In deepest brances of Crenarchaea? Euryarchaea? Archaeal Lipids • Methanogens – Di- and tetra-ethers of glycerol and isoprenoid alcohols – Core mostly archaeol or caldarchaeol – Core sometimes sn-2- or Images removed due to sn-3-hydroxyarchaeol or copyright considerations. macrocyclic archaeol –PMI • Halophiles – Similar to methanogens – Exclusively synthesize bacterioruberin • Marine Crenarchaea Depositional Archaeal Lipids Biological Origin Environment Crocetane methanotrophs? methane seeps? methanogens, PMI (2,6,10,15,19-pentamethylicosane) methanotrophs hypersaline, anoxic Squalane hypersaline? C31-C40 head-to-head isoprenoids Smit & Mushegian • “Lost” enzymes of MVA pathway must exist – Phosphomevalonate kinase (PMK) – Diphosphomevalonate decarboxylase – Isopentenyl diphosphate isomerase (IPPI) Kaneda et al. 2001 Rohdich et al. 2001 Boucher et al. • Isoprenoid biosynthesis of archaea evolved through a combination of processes – Co-option of ancestral enzymes – Modification of enzymatic specificity – Orthologous and non-orthologous gene -

OCEAN/ESS 410 1 Lab 3. the Juan De Fuca Plate Please Write out Your
OCEAN/ESS 410 Lab 3. The Juan de Fuca Plate Please write out your answers on a separate sheet of paper. In this exercise you are going to be looking at the bathymetry of the Juan de Fuca plate region, identifying interesting features and their characteristics, and attempting to interpret what you see. Some of the questions may be difficult for you to answer now (although hopefully not at the end of the Quarter) – part of the objective is to get you to look carefully at the maps and think rather that immediately writing down an answer. The Instructor and TAs will be coming around the lab answering questions. Figure 1 shows the plate boundaries for the Juan de Fuca plate region and you will use this as your road map as you explore the Juan de Fuca plate with the program GeoMapApp, a versatile program that is used by researchers to look at bathymetry and other seafloor data. To run GeoMapApp do the following 1. Start GeoMapApp on the lab computer from the Start menu or from the Desktop Icon if you have one. If it asks you to install a new version you do not have to. 2. Select the default Mercator Base Map (left hand map) 3. Learn to use the “Zoom In”, “Zoom Out” and “Pan the Map” tools. You can use the “Zoom In” tool either by clicking on the map or holding the mouse button down to rubber-band a box of interest. The Overlays menu allows you to add a Distance Scale and Color Bar. -

Archaeology of Eukaryotic DNA Replication
Downloaded from http://cshperspectives.cshlp.org/ on September 25, 2021 - Published by Cold Spring Harbor Laboratory Press Archaeology of Eukaryotic DNA Replication Kira S. Makarova and Eugene V. Koonin National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Bethesda, Maryland 20894 Correspondence: [email protected] Recent advances in the characterization of the archaeal DNA replication system together with comparative genomic analysis have led to the identification of several previously un- characterized archaeal proteins involved in replication and currently reveal a nearly com- plete correspondence between the components of the archaeal and eukaryotic replication machineries. It can be inferred that the archaeal ancestor of eukaryotes and even the last common ancestor of all extant archaea possessed replication machineries that were compa- rable in complexity to the eukaryotic replication system. The eukaryotic replication system encompasses multiple paralogs of ancestral components such that heteromeric complexes in eukaryotes replace archaeal homomeric complexes, apparently along with subfunctionali- zation of the eukaryotic complex subunits. In the archaea, parallel, lineage-specific dupli- cations of many genes encoding replication machinery components are detectable as well; most of these archaeal paralogs remain to be functionally characterized. The archaeal rep- lication system shows remarkable plasticity whereby even some essential components such as DNA polymerase and single-stranded DNA-binding protein are displaced by unrelated proteins with analogous activities in some lineages. ouble-stranded DNA is the molecule that Okazaki fragments (Kornberg and Baker 2005; Dcarries genetic information in all cellular Barry and Bell 2006; Hamdan and Richardson life-forms; thus, replication of this genetic ma- 2009; Hamdan and van Oijen 2010). -

Recent Movements of the Juan De Fuca Plate System
JOURNAL OF GEOPHYSICAL RESEARCH, VOL. 89, NO. B8, PAGES 6980-6994, AUGUST 10, 1984 Recent Movements of the Juan de Fuca Plate System ROBIN RIDDIHOUGH! Earth PhysicsBranch, Pacific GeoscienceCentre, Departmentof Energy, Mines and Resources Sidney,British Columbia Analysis of the magnetic anomalies of the Juan de Fuca plate system allows instantaneouspoles of rotation relative to the Pacific plate to be calculatedfrom 7 Ma to the present.By combiningthese with global solutions for Pacific/America and "absolute" (relative to hot spot) motions, a plate motion sequencecan be constructed.This sequenceshows that both absolute motions and motions relative to America are characterizedby slower velocitieswhere younger and more buoyant material enters the convergencezone: "pivoting subduction."The resistanceprovided by the youngestportion of the Juan de Fuca plate apparently resulted in its detachmentat 4 Ma as the independentExplorer plate. In relation to the hot spot framework, this plate almost immediately began to rotate clockwisearound a pole close to itself such that its translational movement into the mantle virtually ceased.After 4 Ma the remainder of the Juan de Fuca plate adjusted its motion in responseto the fact that the youngest material entering the subductionzone was now to the south. Differencesin seismicityand recent uplift betweennorthern and southernVancouver Island may reflect a distinction in tectonicstyle betweenthe "normal" subductionof the Juan de Fuca plate to the south and a complex "underplating"occurring as the Explorer plate is overriddenby the continent.The history of the Explorer plate may exemplifythe conditionsunder which the self-drivingforces of small subductingplates are overcomeby the influenceof larger, adjacent plates. The recent rapid migration of the absolutepole of rotation of the Juan de Fuca plate toward the plate suggeststhat it, too, may be nearingthis condition. -

The Official Magazine of The
OceTHE OFFICIALa MAGAZINEn ogOF THE OCEANOGRAPHYra SOCIETYphy CITATION Smith, L.M., J.A. Barth, D.S. Kelley, A. Plueddemann, I. Rodero, G.A. Ulses, M.F. Vardaro, and R. Weller. 2018. The Ocean Observatories Initiative. Oceanography 31(1):16–35, https://doi.org/10.5670/oceanog.2018.105. DOI https://doi.org/10.5670/oceanog.2018.105 COPYRIGHT This article has been published in Oceanography, Volume 31, Number 1, a quarterly journal of The Oceanography Society. Copyright 2018 by The Oceanography Society. All rights reserved. USAGE Permission is granted to copy this article for use in teaching and research. Republication, systematic reproduction, or collective redistribution of any portion of this article by photocopy machine, reposting, or other means is permitted only with the approval of The Oceanography Society. Send all correspondence to: [email protected] or The Oceanography Society, PO Box 1931, Rockville, MD 20849-1931, USA. DOWNLOADED FROM HTTP://TOS.ORG/OCEANOGRAPHY SPECIAL ISSUE ON THE OCEAN OBSERVATORIES INITIATIVE The Ocean Observatories Initiative By Leslie M. Smith, John A. Barth, Deborah S. Kelley, Al Plueddemann, Ivan Rodero, Greg A. Ulses, Michael F. Vardaro, and Robert Weller ABSTRACT. The Ocean Observatories Initiative (OOI) is an integrated suite of instrumentation used in the OOI. The instrumented platforms and discrete instruments that measure physical, chemical, third section outlines data flow from geological, and biological properties from the seafloor to the sea surface. The OOI ocean platforms and instrumentation to provides data to address large-scale scientific challenges such as coastal ocean dynamics, users and discusses quality control pro- climate and ecosystem health, the global carbon cycle, and linkages among seafloor cedures. -
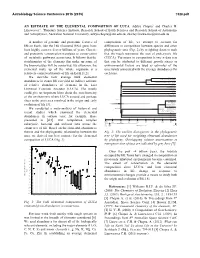
An Estimate of the Elemental Composition of Luca
Astrobiology Science Conference 2015 (2015) 7328.pdf AN ESTIMATE OF THE ELEMENTAL COMPOSITION OF LUCA. Aditya Chopra1 and Charles H. Lineweaver1, 1Planetary Science Institute, Research School of Earth Sciences and Research School of Astronomy and Astrophysics, Australian National University, [email protected], [email protected] A number of genomic and proteomic features of composition of life, we attempt to account for life on Earth, like the 16S ribosomal RNA gene, have differences in composition between species and other been highly conserved over billions of years. Genetic phylogenetic taxa (Fig. 2) by weighting datasets such and proteomic conservation translates to conservation that the result represents the root of prokaryotic life of metabolic pathways across taxa. It follows that the (LUCA). Variations in composition between data sets stoichiometry of the elements that make up some of that can be attributed to different growth stages or the biomolecules will be conserved. By extension, the environmental factors are used as estimates of the elemental make up of the whole organism is a uncertainty associated with the average abundances for relatively conserved feature of life on Earth [1,2]. each taxa. We describe how average bulk elemental Euryarchaeota 1a Methanococci, Methanobacteria, Methanopyri 7 Euryarchaeota 1b abundances in extant life can yield an indirect estimate 6 Thermoplasmata, Methanomicrobia, Halobacteria, Archaeoglobi Euryarchaeota 2 4 Thermococci of relative abundances of elements in the Last Crenarchaeota Sulfolobus, Thermoproteus ? Thaumarchaeota Universal Common Ancestor (LUCA). The results Cenarchaeum ? Korarchaeota ARMAN could give us important hints about the stoichiometry ? 2 Archaeal Richmond Mine Acidophilic Nanoorganisms Nanoarchaeota of the environment where LUCA existed and perhaps Archaea Terrabacteria clues to the processes involved in the origin and early Actinobacteria, Deinococcus-Thermus, 1 Cyanobacteria, Life Chloroflexi, 9 evolution of life [3]. -

Universidad Politécnica De Madrid Escuela Técnica Superior De Ingeniería Agronómica Alimentaria Y De Biosistemas Departamento De Biotecnología-Biología Vegetal
UNIVERSIDAD POLITÉCNICA DE MADRID ESCUELA TÉCNICA SUPERIOR DE INGENIERÍA AGRONÓMICA ALIMENTARIA Y DE BIOSISTEMAS DEPARTAMENTO DE BIOTECNOLOGÍA-BIOLOGÍA VEGETAL TESIS DOCTORAL Caracterización de una nueva proteína hipertermófila NifB y su papel en el mecanismo de síntesis de NifB-co, precursor del grupo metálico de FeMo-co Author: Alessandro Scandurra Director: Prof. Luis Manuel Rubio Herrero Co-director: Dr. Simon Jean Marius Arragain i ii UNIVERSIDAD POLITÉCNICA DE MADRID ESCUELA TÉCNICA SUPERIOR DE INGENIERÍA AGRONÓMICA ALIMENTARIA Y DE BIOSISTEMAS DEPARTAMENTO DE BIOTECNOLOGÍA-BIOLOGÍA VEGETAL Memoria presentada por D. Alessandro Scandurra para optar al grado de Doctor Director Dr. Luis Manuel Rubio Herrero Profesor Titular UPM Madrid, 2017 © This copy oF the thesis has been supplied on condition that anyone who consults it is understood to recognize that its copyright rest with the author and that no quotation From the thesis, or any inFormation derived therefrom may be published without the author’s prior, written consent. I II AcKnowledgments As each journey, also this one has an end (Thanks goodness). It was a long trip, with several changes oF direction. Like For many other people, the PhD adventure was not easy, but I have to admit that I was lucky enough to meet really unique people, that became special Friendships. First oF all, I want to thank my Family, especially my mother and my Father, who always push me to go Forward in my life and to Follow my dreams, ready to pay the sacrifice oF the distance. This expirience has helped me to understand how easy is to be a son and how challenging is to be a parent.