Xu Jiang Last Update: Wednesday 14Th August, 2019
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

TRIAL INNOVATION NETWORK Team Roster
TRIAL INNOVATION NETWORK Team Roster Trial Innovation Center – Duke University/Vanderbilt University Danny Benjamin Michael DeBaun Julie Ozier Principal Investigator Vanderbilt Investigator Representative C-IRB Lead Duke University Vanderbilt University Vanderbilt University [email protected] [email protected] [email protected] Gordon Bernard Jennifer Dix JoAnna Pomerantz Co-Principal Investigator Admin Support- Website Sr. Associate Contracts Management Vanderbilt University Vanderbilt University Duke University [email protected] [email protected] [email protected] Lori Poole Julia Dunagan Renee Pridgen Lead Program Manager Admin Support- Website Director, Clinical Operations Duke University Vanderbilt University Duke University [email protected] [email protected] [email protected] Terri Edwards Aimee Edgeworth Jill Pulley Admin Lead Admin Support Executive Director Vanderbilt University Vanderbilt University Vanderbilt University [email protected] [email protected] [email protected] Rebecca Abel Shelby Epps Libby Salberg C-IRB Lead Admin Support-Master Agreements Master Agreements Project Lead Vanderbilt University Vanderbilt University Vanderbilt University [email protected] [email protected] [email protected] Leslie Amos Davera Gabriel Emily Sheffer Project Lead Senior Informaticist Admin Support- Central IRB Duke University Duke University Vanderbilt University [email protected] [email protected] -

View from the Chair
View from the Chair This past summer we initiated a major new program for John Imbrie research experiences for undergraduates (REUs), Professor of Mathematics/Chair combining Ken Ono's long-running program in number theory with a new program in geometry and topology, I write at the conclusion of an extraordinary year for our supported by the RTG grant. Despite the challenges of work- department. As 2020 began, we were gearing up to host the ing remotely, the program was very successful -- see the AMS sectional, which was to start on March 12. Early article below by Ken Ono and Tom Mark. Other indications were that large gatherings were a major highlights of this Virginia Math Bulletin include an article on contributor to the spread of the pandemic, so we hastily our new bridge program, by David Sherman (our new cancelled the event, which was to bring 700 participants to Director of Diversity, Equity, and Inclusion). Our Bridge to Charlottesville. Soon, the university cancelled classes and the Doctorate currently supports three students were abruptly sent post-baccalaureate students as they home. Faculty members and prepare for entry into a Ph.D. program. graduate students were tasked with quickly finding Sadly, it was impossible to conduct the a way to hold classes online. Gordon Keller math majors dinner. We The learning curve was had planned to host UVa graduate steep, but we came together and McShane Prize winner Adrew Booker, to reinvent our modes of who was in the news for work leading to teaching. Now we reach the the long-sought integer solutions to conclusion of a semester x3 + y3 + z3 = 33 and x3 + y3 + z3 = 42. -

Proposal Description
updated 2/8/21 Comparative Thought and Literature Johns Hopkins University 410.516.2367 (office) 3400 N. Charles St, Gilman 226 [email protected] Baltimore, MD 21218 https://lisasiraganian.com Lisa Michele Siraganian Academic Positions James R. Herbert Boone Chair in Comparative Thought and Literature and Associate Professor (with tenure). July 1, 2019–. Department of Comparative Thought and Literature. Johns Hopkins University (Baltimore, MD). Associate Professor (with tenure). 2012–2019. Department of English. Assistant Professor. 2005–12. Department of English. Southern Methodist University (Dallas, TX). Administrative Positions Chair, Department of Comparative Thought and Literature. The Johns Hopkins University (Baltimore, MD). July 1, 2019—. Ruth Collins Altshuler Director, Dedman College Interdisciplinary Institute. Southern Methodist University (Dallas, TX). 2018–2019. Associate Director, Dedman College Interdisciplinary Institute. Southern Methodist University (Dallas, TX). 2013–2015. Education Dedman School of Law, Southern Methodist University. J.D. May 2019 (Cum Laude. Evening program. One year of coursework at Harvard University) Johns Hopkins University. English and American Literature. M.A. 2000, Ph.D. 2004 (George E. Owen Dean’s Fellowship. Dean’s Teaching Fellowship) Oxford University. Faculty of English Language and Literature. B.A. 1997 (First Class Honors, I. Exeter College Fitzgerald Prize) Williams College. Honors in English Literature. B.A.1995 (Summa cum laude. Elizabeth Shumway Prize in English. Phi Beta Kappa junior year) Siraganian -- 2 National and Residential Fellowships New Directions Fellowship, Andrew W. Mellon Foundation. 2015-2018. ($222,000.00 award). American Council for Learned Societies [ACLS] Fellowship. 2015-2016. American Academy of Arts and Sciences (Cambridge, Massachusetts). Visiting Scholar Residential Fellowship. 2011-2012. -

Duke Staff Handbook
DUKE STAFF HANDBOOK “…itisuptoustoset anexampleofhowa communityofpeople workingtogethertoward acommonpurpose canrealizeoutrageous ambitions.” VincentE.Price President Checkout ,~,_ UKE forthelatestnews,resources&conversation Print: Working@Dukepublication Online:working.duke.edu Facebook:facebook.com/workingatduke 0 Twitter: twitter.com/workingatduke WELCOME TO DUKE As President, it is my pleasure to welcome you to Duke! Duke’s employees are among the university’s greatest strengths. When you work here, you don’t just do the job and go home. You’re part of the campus community. You share in the university’s successes and help define its identity. Former president Terry Sanford perhaps put it best: every person who works for Duke is important to Duke; they are all Duke University People. As a new Duke University Person, you should take some time to familiarize yourself with this handbook, which is intended to help you establish a successful working relationship with the Duke community. It outlines the many resources and opportunities that are available to you as an employee, and it should help you understand what Duke expects from you as a staff member and what you should expect from Duke. You can find answers to additional questions by reviewing the Duke Human Resources Policy Manual (hr.duke.edu/policies) or by speaking with your supervisor. I also invite you to share in the responsibility of shaping the university’s identity and guiding it toward a more inclusive future. Duke is about much more than what happens in the classroom or the lab; it is up to us to set an example of how a community of people working together toward a common purpose can realize outrageous ambitions. -

Heather Lynch - Graduate Student in Molecular Genetics and Microbiology Duke University • Richard W
SERCEB Policy, Ethics and Law Core: Biosecurity and Dual-use Education Activities Southeastern Regional Center of Excellence for Emerging Infections and Biodefense (UNC, U Fl, UAB, Emory, Vanderbilt, Duke & 20+ affiliated groups) www.serceb.org/pel/ Members and staff Allison Chamberlain, MS Duke University (position formerly held by Megan Davidson) Nikki M. Vangsnes, Duke University Robert M. Cook-Deegan, MD Duke University Ross McKinney, MD, Duke University Ruth Berkelman, MD, Emory University Arri Eisen, PhD, Emory University LouAnn C. Burnett, MS, CBSP, Vanderbilt University Elizabeth Heitman, PhD, Vanderbilt University Nancy M P King, JD, Wake Forest University Rebecca Walker, Ph.D., University of North Carolina – Chapel Hill Paul Gulig, PhD, University of Florida James Thomas, PhD, MPH, UNC School of Public Health Samuel Tilden, MD, JD, LLM, University of Alabama Mechanisms Talk to investigators and staff Meet with IRBs, IBCs, IUCACs, etc. Online education module Biosecurity/dual-use education panels (Duke 2006; Emory 2008) Policy Engagement (NSABB, NRC, ASM, etc.) International Engagement (South Africa, Poland, Japan, Hungary) Presence at relevant conferences (ABSA, ASM Biodefense, ICEID) White papers (BWC, role of scientists, IRB review snags, animals in research, etc.) Active dual-use review of SERCEB-funded projects through Steering Committee Summary of PEL experience Science 316 (8 June): 1432-33, 2007. SERCEB PEL Dual-use Educational Module Developed as a tool to teach scientists (PIs, post-docs, lab -

March 2019 CURRICULUM VITAE DENNIS TAO YANG PERSONAL INFORMATION Work Address
March 2019 CURRICULUM VITAE DENNIS TAO YANG PERSONAL INFORMATION Work Address: Darden School of Business University of Virginia (UVA) Charlottesville, VA 22906, USA Phone: (434) 9240906 Email: [email protected] Date and Place of Birth: February 1, 1966; Beijing, China Marital Status: Married; two children Citizenship: United States of America EDUCATION 1994 Ph.D., Economics, University of Chicago 1987 B.A., Economics, Magna Cum Laude, University of California, Los Angeles 1984 International Baccalaureate, United World College, Trieste, Italy FIELDS OF INTEREST Economic Development and Growth Labor and Demographic Economics Economics of China and Transition PROFESSIONAL EXPERIENCE Academic Positions: 2013 Dale S. Coenen Free Enterprise Professor, Darden School of Business, UVA 2012 Professor, Darden School of Business, UVA 201316 Chang Jiang Scholar Professorship, Antai College of Economics and Management, Shanghai Jiao Tong University, China 200712 Professor, Department of Economics, Chinese University of Hong Kong (CUHK) 200607 Professor, Department of Economics, Virginia Polytechnic Institute and State University (Virginia Tech); 20012005, Associate Professor 19942001 Assistant Professor, Department of Economics, Duke University 2012 Senior Fellow, Economic Research Center, Hong Kong Institute of Asia-Pacific Studies, CUHK 2010 Research Fellow, Institute for the Study of Labor (IZA), Germany 200514 Senior Fellow, Center for China in the World Economy, Tsinghua University 200713 Senior Fellow, China Center for Public Finance, -

Duke University 2002-2003
bulletin of Duke University 2002-2003 The Fuqua School of Business University’s Mission Statement James B. Duke’s founding Indenture of Duke University directed the members of the University to “provide real leadership in the educational world” by choosing indi- viduals of “outstanding character, ability and vision” to serve as its officers, trustees and faculty; by carefully selecting students of “character, determination and application;” and by pursuing those areas of teaching and scholarship that would “most help to de- velop our resources, increase our wisdom, and promote human happiness.” To these ends, the mission of Duke University is to provide a superior liberal educa- tion to undergraduate students, attending not only to their intellectual growth but also to their development as adults committed to high ethical standards and full participa- tion as leaders in their communities; to prepare future members of the learned profes- sions for lives of skilled and ethical service by providing excellent graduate and professional education; to advance the frontiers of knowledge and contribute boldly to the international community of scholarship; to promote an intellectual environment built on a commitment to free and open inquiry; to help those who suffer, cure disease and promote health, through sophisticated medical research and thoughtful patient care; to provide wide ranging educational opportunities, on and beyond our campuses, for traditional students, active professionals and life-long learners using the power of in- formation technologies; and to promote a deep appreciation for the range of human dif- ference and potential, a sense of the obligations and rewards of citizenship, and a commitment to learning, freedom and truth. -

Duke Office of Audit, Risk, and Compliance
DUKE UNIVERSITY Statement of Ethical Principles and Code of Conduct I. STATEMENT OF PURPOSE Duke University is an institution of higher education dedicated to excellence in education, research and patient care. Throughout its history, Duke has been dedicated to upholding the highest ethical standards in its students, faculty and staff. As stated in the Duke University Mission Statement, “by pursuing these objectives with vision and integrity, Duke University seeks to engage the mind, elevate the spirit, and stimulate the best effort of all who are associated with the university.” II. SCOPE This document serves as a statement of principles and responsibilities for the full Duke community. Members of the Duke University, including Duke University Health System (DUHS), community include trustees, senior officials, faculty, staff, students, student employees, student leaders and university-authorized volunteers acting on behalf of Duke. III. STATEMENT OF ETHICAL PRINCIPLES This Statement of Ethical Principles sets forth overarching ethical principles to which members of the Duke University community are expected to adhere. These ethical principles are intended to provide a foundation for conduct in support of the university's mission. This Statement of Ethical Principles should be used as a general guide in making ethical decisions that affect the Duke University community. These Principles also can be found in various forms in separate codes of conduct, policies and procedures of the Duke University community, including the Duke Guiding Principles, the DUHS Values and the Duke Community Standard. The university community is diverse – in race, ethnicity, gender, sexual orientation, background, age, religion, abilities and in many other ways. -

Silverman Cv.Pdf
Justin Silverman Contact [email protected] justin-silverman.com Information statsathome.com Research Bayesian statistics, time series analysis, multivariate analysis, sequence count data, Interests (e.g., microbiome surveys, gene expression studies, single cell RNA-seq), epidemiology, compositional data analysis, decision theory. Employment Pennsylvania State University 2020- Assistant Professor, College of Information Science and Technology (Primary) Assistant Professor, Department of Medicine (Secondary) Education MD and PhD, Duke University 2012-2020 PhD in Computational Biology and Bioinformatics Thesis: Bayesian Multivariate Count Models for the Design and Analysis of Microbiome Studies Mentors: Dr. Lawrence David and Dr. Sayan Mukherjee Research Technician, Johns Hopkins University 2011-2012 Mentor: Dr. Nina Markovic Bachelors of Science, Johns Hopkins University 2007-2011 Double Major in Physics and Biophysics, Minor in Mathematics Publications [∗ denotes equal contribution] Silverman JD∗, Hupert N, Washburne AD∗. Using influenza surveillance networks to estimate state-specific case detection-rates and forecast SARS-CoV-2 spread in the United States. (2020) medRxiv, DOI: 10.1101/2020.04.01.20050542 Morton J∗, Marotz C∗, Washburne A, Silverman JD, Edlund A, Zaramela L, Zengler K, Knight R. (2019) Establishing microbial measurement standards with reference frames. Nature Communications, 2719; DOI: 0.1038/s41467-019-10656-5 Villa MM∗, Bloom RJ∗, Silverman JD, Durand HK, Jiang S, Wu A, Huang S, You L, David LA. (2019) High-throughput isolation and culture of human gut bacteria with droplet microfluidics. bioRxiv 630822; DOI:10.1101/630822 Silverman JD, Bloom RJ, Jiang S, Durand HK, Mukherjee S, David LA. (2019) Mea- suring and Mitigating PCR Bias in Microbiome Data. bioRxiv 604025; DOI:10.1101/604025 Silverman JD, Roche K, Holmes ZC, David LA, Mukherjee S. -
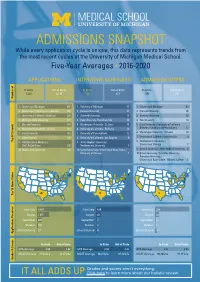
5-Year Admissions Snapshot
ADMISSIONS SNAPSHOT While every application cycle is unique, this data represents trends from the most recent cycles at the University of Michigan Medical School. Five-Year Averages • 2016-2020 APPLICATIONS INTERVIEWS SCHEDULED ADMISSION OFFERS In State Out of State In State Out of State In State Out of State 1,222 6,130 123 372 100 281 Number of 1. University of Michigan 607 1. University of Michigan 87 1. University of Michigan 69 2. University of California - Los Angeles 269 2. Harvard University 27 2. Harvard University 25 3. University of California - Berkeley 239 3. Stanford University 18 3. Stanford University 16 4. Michigan State University 177 4. Duke University /Yale University 16 4. Yale University 14 5. Harvard University 134 5. Washington University - St. Louis 15 5. Duke University / University of California - 6. Washington University - St. Louis 118 6. University of California - Berkeley 14 Berkeley / University of Pennsylvania 12 7. Duke University 110 7. University of Pennsylvania 12 6. Washington University - St. Louis 10 8. Johns Hopkins 107 8. University of California - Los Angeles 9 7. University of California - Los Angeles 9 Top 10 Schools Top 9. U of Wisconsin-Madison / 9. Johns Hopkins University / 8. Northwestern University / U of CA-San Diego 105 Northwestern University 8 University of Chicago 7 10. Wayne State University 103 10. Cornell University / University of Notre Dame / 9. Cornell University / Johns Hopkins University 6 University of Chicago 7 10. Brown University / Columbia University / Princeton University -

Appendix P: Policies Related to Research
APPENDIX P: POLICIES RELATED TO RESEARCH In this Appendix: APPENDIX P: POLICIES RELATED TO RESEARCH ..................................................................................... 1 Protecting Human Subjects in Nonmedical Research ......................................................................................... 2 Policy on Inventions, Patents, and Technology Transfer .................................................................................... 7 Interpretation of the Policy on Inventions, Patents, and Technology Transfer Reaffirming the Rights of Undergraduate Students .................................................................................................................................... 13 Duke University Policy on Intellectual Property Rights ................................................................................... 14 Interpretations of the Policy on Intellectual Property Rights as it Relates to Online Courseware .................... 19 Duke University Copyright Guidelines for Electronic Course Content ............................................................ 21 Checklist for Fair Use Analysis ........................................................................................................................ 22 Patent Agreement .............................................................................................................................................. 24 University-Industry Guidelines ........................................................................................................................ -

Aaron D. Franklin 130 Hudson Hall, Box 90291 Durham, NC 27708 Curriculum Vita Phone: (919) 681-9471 Email: [email protected] Website: Franklin.Pratt.Duke.Edu
Address: Duke University Aaron D. Franklin 130 Hudson Hall, Box 90291 Durham, NC 27708 Curriculum Vita Phone: (919) 681-9471 Email: [email protected] Website: franklin.pratt.duke.edu Education Ph.D. Electrical Engineering Purdue University 2008 West Lafayette, IN B.S. Electrical Engineering Arizona State University 2004 Minor: Communication Tempe, AZ Honors and Awards • Dean’s Award for Excellence in Mentoring 2021 • Capers & Marion McDonald Award for Excellence in Teaching and Research 2020 • Best Poster Award (advisor) at Device Research Conference (DRC) 2020 • Bass Chair & Fellow of Bass Society at Duke 2018 • Best Poster Award (advisor) at MRS Fall Meeting 2018 • Best Paper Award (advisor) at international IEEE Sensors conference 2017 • 2nd Place at MRS iMatSci Innovation Showcase for Tire Tread Wear Sensor Technology 2017 • Best Poster Award (advisor) at Device Research Conference (DRC) 2017 • IBM Labyrinth Award for Invention Accomplishments 2014 • IBM Outstanding Technical Achievement Award 2014 • IBM Research Outstanding Contributor Award 2013 • GOMAC Technology Conference #1 Outstanding Paper Award 2012 • IBM Invention Achievement Awards 2009, 2011, 2012, 2013 • GOMAC Technology Conference Meritorious Paper Award 2010 • National Science Foundation (NSF) Graduate Research Fellowship (GRF) 2005 – 2008 • First place poster prize and NSF travel scholarship for Nano and Giga Conference 2007 • Materials Research Society (MRS) Graduate Student Silver Award 2006 • NASA Institute for Nanoelectronics and Computing (INaC) Fellowship