First Report About the Identification and Preliminary Analysis of a Partial Sequence of Dihydropyrimidine Dehydrogenase (NADP+)
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

A Proteomic Approach to Uncover Neuroprotective Mechanisms of Oleocanthal Against Oxidative Stress
International Journal of Molecular Sciences Article A Proteomic Approach to Uncover Neuroprotective Mechanisms of Oleocanthal against Oxidative Stress Laura Giusti 1,†, Cristina Angeloni 2,†, Maria Cristina Barbalace 3, Serena Lacerenza 4, Federica Ciregia 5, Maurizio Ronci 6 ID , Andrea Urbani 7, Clementina Manera 4, Maria Digiacomo 4 ID , Marco Macchia 4, Maria Rosa Mazzoni 4, Antonio Lucacchini 1 ID and Silvana Hrelia 3,* ID 1 Department of Clinical and Experimental Medicine, University of Pisa, 56126 Pisa, Italy; [email protected] (L.G.); [email protected] (A.L.) 2 School of Pharmacy, University of Camerino, 62032 Camerino, Italy; [email protected] 3 Department for Life Quality Studies, Alma Mater Studiorum, University of Bologna, 47921 Rimini, Italy; [email protected] 4 Department of Pharmacy, University of Pisa, 56126 Pisa, Italy; [email protected] (S.L.); [email protected] (C.M.); [email protected] (M.D.); [email protected] (M.M.); [email protected] (M.R.M.) 5 Department of Rheumatology, GIGA Research, Centre Hospitalier Universitaire (CHU) de Liège, University of Liège, 4000 Liège, Belgium; [email protected] 6 Department of Medical, Oral and Biotechnological Sciences, University G. d’Annunzio of Chieti-Pescara, 65127 Pescara, Italy; [email protected] 7 Institute of Biochemistry and Clinical Biochemistry, Catholic University, 00198 Rome, Italy; [email protected] * Correspondence: [email protected]; Tel.: +39-051-209-1235 † These authors contributed equally to this work. Received: 3 July 2018; Accepted: 1 August 2018; Published: 8 August 2018 Abstract: Neurodegenerative diseases represent a heterogeneous group of disorders that share common features like abnormal protein aggregation, perturbed Ca2+ homeostasis, excitotoxicity, impairment of mitochondrial functions, apoptosis, inflammation, and oxidative stress. -

Supplementary Materials
Supplementary Materials COMPARATIVE ANALYSIS OF THE TRANSCRIPTOME, PROTEOME AND miRNA PROFILE OF KUPFFER CELLS AND MONOCYTES Andrey Elchaninov1,3*, Anastasiya Lokhonina1,3, Maria Nikitina2, Polina Vishnyakova1,3, Andrey Makarov1, Irina Arutyunyan1, Anastasiya Poltavets1, Evgeniya Kananykhina2, Sergey Kovalchuk4, Evgeny Karpulevich5,6, Galina Bolshakova2, Gennady Sukhikh1, Timur Fatkhudinov2,3 1 Laboratory of Regenerative Medicine, National Medical Research Center for Obstetrics, Gynecology and Perinatology Named after Academician V.I. Kulakov of Ministry of Healthcare of Russian Federation, Moscow, Russia 2 Laboratory of Growth and Development, Scientific Research Institute of Human Morphology, Moscow, Russia 3 Histology Department, Medical Institute, Peoples' Friendship University of Russia, Moscow, Russia 4 Laboratory of Bioinformatic methods for Combinatorial Chemistry and Biology, Shemyakin-Ovchinnikov Institute of Bioorganic Chemistry of the Russian Academy of Sciences, Moscow, Russia 5 Information Systems Department, Ivannikov Institute for System Programming of the Russian Academy of Sciences, Moscow, Russia 6 Genome Engineering Laboratory, Moscow Institute of Physics and Technology, Dolgoprudny, Moscow Region, Russia Figure S1. Flow cytometry analysis of unsorted blood sample. Representative forward, side scattering and histogram are shown. The proportions of negative cells were determined in relation to the isotype controls. The percentages of positive cells are indicated. The blue curve corresponds to the isotype control. Figure S2. Flow cytometry analysis of unsorted liver stromal cells. Representative forward, side scattering and histogram are shown. The proportions of negative cells were determined in relation to the isotype controls. The percentages of positive cells are indicated. The blue curve corresponds to the isotype control. Figure S3. MiRNAs expression analysis in monocytes and Kupffer cells. Full-length of heatmaps are presented. -

Dihydropyrimidinase Deficiency
Dihydropyrimidinase deficiency Description Dihydropyrimidinase deficiency is a disorder that can cause neurological and gastrointestinal problems in some affected individuals. Other people with dihydropyrimidinase deficiency have no signs or symptoms related to the disorder, and in these individuals the condition can be diagnosed only by laboratory testing. The neurological abnormalities that occur most often in people with dihydropyrimidinase deficiency are intellectual disability, seizures, and weak muscle tone (hypotonia). An abnormally small head size (microcephaly) and autistic behaviors that affect communication and social interaction also occur in some individuals with this condition. Gastrointestinal problems that occur in dihydropyrimidinase deficiency include backflow of acidic stomach contents into the esophagus (gastroesophageal reflux) and recurrent episodes of vomiting (cyclic vomiting). Affected individuals can also have deterioration ( atrophy) of the small, finger-like projections (villi) that line the small intestine and provide a large surface area with which to absorb nutrients. This condition, called villous atrophy, can lead to difficulty absorbing nutrients from foods (malabsorption), resulting in a failure to grow and gain weight at the expected rate (failure to thrive). People with dihydropyrimidinase deficiency, including those who otherwise exhibit no symptoms, may be vulnerable to severe, potentially life-threatening toxic reactions to certain drugs called fluoropyrimidines that are used to treat cancer. Common examples of these drugs are 5-fluorouracil and capecitabine. These drugs may not be broken down efficiently and can build up to toxic levels in the body (fluoropyrimidine toxicity), leading to drug reactions including gastrointestinal problems, blood abnormalities, and other signs and symptoms. Frequency Dihydropyrimidinase deficiency is thought to be a rare disorder. -

Genetic Factors Influencing Pyrimidine- Antagonist Chemotherapy
The Pharmacogenomics Journal (2005) 5, 226–243 & 2005 Nature Publishing Group All rights reserved 1470-269X/05 $30.00 www.nature.com/tpj REVIEW Genetic factors influencing Pyrimidine- antagonist chemotherapy JG Maring1 ABSTRACT 2 Pyrimidine antagonists, for example, 5-fluorouracil (5-FU), cytarabine (ara-C) HJM Groen and gemcitabine (dFdC), are widely used in chemotherapy regimes for 2 FM Wachters colorectal, breast, head and neck, non-small-cell lung cancer, pancreatic DRA Uges3 cancer and leukaemias. Extensive metabolism is a prerequisite for conversion EGE de Vries4 of these pyrimidine prodrugs into active compounds. Interindividual variation in the activity of metabolising enzymes can affect the extent of 1Department of Pharmacy, Diaconessen Hospital prodrug activation and, as a result, act on the efficacy of chemotherapy Meppel & Bethesda Hospital Hoogeveen, Meppel, treatment. Genetic factors at least partly explain interindividual variation in 2 The Netherlands; Department of Pulmonary antitumour efficacy and toxicity of pyrimidine antagonists. In this review, Diseases, University of Groningen & University Medical Center Groningen, Groningen, The proteins relevant for the efficacy and toxicity of pyrimidine antagonists will Netherlands; 3Department of Pharmacy, be summarised. In addition, the role of germline polymorphisms, tumour- University of Groningen & University Medical specific somatic mutations and protein expression levels in the metabolic Center Groningen, Groningen, The Netherlands; pathways and clinical pharmacology -

Genome-Wide Transcriptional Analysis of Carboplatin Response in Chemosensitive and Chemoresistant Ovarian Cancer Cells
1605 Genome-wide transcriptional analysis of carboplatin response in chemosensitive and chemoresistant ovarian cancer cells David Peters,1,2 John Freund,2 and Robert L. Ochs2 variety of disease processes, including cancer. Patterns of global gene expression can reveal the molecular pathways 1 Department of Pharmacology and Therapeutics, University of relevant to the disease process and identify potential new Liverpool, United Kingdom and 2Precision Therapeutics, Inc., Pittsburgh, Pennsylvania therapeutic targets. The use of this technology for the molecular classification of cancer was recently shown with the identification of an expression profile that was Abstract predictive of patient outcome for B-cell lymphoma (1). We have recently described an ex vivo chemoresponse In addition, this study showed that histologically similar assay for determining chemosensitivity in primary cultures tumors can be differentiated based on their gene expression of human tumors. In this study, we have extended these profiles. Ultimately, these unique patterns of gene expres- experiments in an effort to correlate chemoresponse data sion may be used as guidelines to direct different modes of with gene expression patterns at the level of transcription. therapy. Primary cultures of cells derived from ovarian carcinomas Although it is widely recognized that patients with the of individual patients (n = 6) were characterized using same histologic stage and grade of cancer respond to the ChemoFx assay and classified as either carboplatin therapies differently, few clinical tests can predict indi- sensitive (n = 3) or resistant (n = 3). Three representa- vidual patient responses. The next great challenge will be tive cultures of cells from each individual tumor were then to use the power of post-genomic technology, including subjected to Affymetrix gene chip analysis (n = 18) using microarray analyses, to correlate gene expression patterns U95A human gene chip arrays. -

Integrative Analysis of Disease Signatures Shows Inflammation Disrupts Juvenile Experience-Dependent Cortical Plasticity
New Research Development Integrative Analysis of Disease Signatures Shows Inflammation Disrupts Juvenile Experience- Dependent Cortical Plasticity Milo R. Smith1,2,3,4,5,6,7,8, Poromendro Burman1,3,4,5,8, Masato Sadahiro1,3,4,5,6,8, Brian A. Kidd,2,7 Joel T. Dudley,2,7 and Hirofumi Morishita1,3,4,5,8 DOI:http://dx.doi.org/10.1523/ENEURO.0240-16.2016 1Department of Neuroscience, Icahn School of Medicine at Mount Sinai, New York, New York 10029, 2Department of Genetics and Genomic Sciences, Icahn School of Medicine at Mount Sinai, New York, New York 10029, 3Department of Psychiatry, Icahn School of Medicine at Mount Sinai, New York, New York 10029, 4Department of Ophthalmology, Icahn School of Medicine at Mount Sinai, New York, New York 10029, 5Mindich Child Health and Development Institute, Icahn School of Medicine at Mount Sinai, New York, New York 10029, 6Graduate School of Biomedical Sciences, Icahn School of Medicine at Mount Sinai, New York, New York 10029, 7Icahn Institute for Genomics and Multiscale Biology, Icahn School of Medicine at Mount Sinai, New York, New York 10029, and 8Friedman Brain Institute, Icahn School of Medicine at Mount Sinai, New York, New York 10029 Visual Abstract Throughout childhood and adolescence, periods of heightened neuroplasticity are critical for the development of healthy brain function and behavior. Given the high prevalence of neurodevelopmental disorders, such as autism, identifying disruptors of developmental plasticity represents an essential step for developing strategies for prevention and intervention. Applying a novel computational approach that systematically assessed connections between 436 transcriptional signatures of disease and multiple signatures of neuroplasticity, we identified inflammation as a common pathological process central to a diverse set of diseases predicted to dysregulate Significance Statement During childhood and adolescence, heightened neuroplasticity allows the brain to reorganize and adapt to its environment. -

O O2 Enzymes Available from Sigma Enzymes Available from Sigma
COO 2.7.1.15 Ribokinase OXIDOREDUCTASES CONH2 COO 2.7.1.16 Ribulokinase 1.1.1.1 Alcohol dehydrogenase BLOOD GROUP + O O + O O 1.1.1.3 Homoserine dehydrogenase HYALURONIC ACID DERMATAN ALGINATES O-ANTIGENS STARCH GLYCOGEN CH COO N COO 2.7.1.17 Xylulokinase P GLYCOPROTEINS SUBSTANCES 2 OH N + COO 1.1.1.8 Glycerol-3-phosphate dehydrogenase Ribose -O - P - O - P - O- Adenosine(P) Ribose - O - P - O - P - O -Adenosine NICOTINATE 2.7.1.19 Phosphoribulokinase GANGLIOSIDES PEPTIDO- CH OH CH OH N 1 + COO 1.1.1.9 D-Xylulose reductase 2 2 NH .2.1 2.7.1.24 Dephospho-CoA kinase O CHITIN CHONDROITIN PECTIN INULIN CELLULOSE O O NH O O O O Ribose- P 2.4 N N RP 1.1.1.10 l-Xylulose reductase MUCINS GLYCAN 6.3.5.1 2.7.7.18 2.7.1.25 Adenylylsulfate kinase CH2OH HO Indoleacetate Indoxyl + 1.1.1.14 l-Iditol dehydrogenase L O O O Desamino-NAD Nicotinate- Quinolinate- A 2.7.1.28 Triokinase O O 1.1.1.132 HO (Auxin) NAD(P) 6.3.1.5 2.4.2.19 1.1.1.19 Glucuronate reductase CHOH - 2.4.1.68 CH3 OH OH OH nucleotide 2.7.1.30 Glycerol kinase Y - COO nucleotide 2.7.1.31 Glycerate kinase 1.1.1.21 Aldehyde reductase AcNH CHOH COO 6.3.2.7-10 2.4.1.69 O 1.2.3.7 2.4.2.19 R OPPT OH OH + 1.1.1.22 UDPglucose dehydrogenase 2.4.99.7 HO O OPPU HO 2.7.1.32 Choline kinase S CH2OH 6.3.2.13 OH OPPU CH HO CH2CH(NH3)COO HO CH CH NH HO CH2CH2NHCOCH3 CH O CH CH NHCOCH COO 1.1.1.23 Histidinol dehydrogenase OPC 2.4.1.17 3 2.4.1.29 CH CHO 2 2 2 3 2 2 3 O 2.7.1.33 Pantothenate kinase CH3CH NHAC OH OH OH LACTOSE 2 COO 1.1.1.25 Shikimate dehydrogenase A HO HO OPPG CH OH 2.7.1.34 Pantetheine kinase UDP- TDP-Rhamnose 2 NH NH NH NH N M 2.7.1.36 Mevalonate kinase 1.1.1.27 Lactate dehydrogenase HO COO- GDP- 2.4.1.21 O NH NH 4.1.1.28 2.3.1.5 2.1.1.4 1.1.1.29 Glycerate dehydrogenase C UDP-N-Ac-Muramate Iduronate OH 2.4.1.1 2.4.1.11 HO 5-Hydroxy- 5-Hydroxytryptamine N-Acetyl-serotonin N-Acetyl-5-O-methyl-serotonin Quinolinate 2.7.1.39 Homoserine kinase Mannuronate CH3 etc. -

Potential Widespread Denitrosylation of Brain Proteins Following Prolonged Restraint: Proposed Links Between Stress and Central Nervous System Disease
Metabolic Brain Disease (2019) 34:183–189 https://doi.org/10.1007/s11011-018-0340-1 ORIGINAL ARTICLE Potential widespread denitrosylation of brain proteins following prolonged restraint: proposed links between stress and central nervous system disease Timothy D. Foley1 & Kari S. Koval1 & Alexandria G. Gallagher1 & Stefan H. Olsen1 Received: 9 September 2018 /Accepted: 4 November 2018 /Published online: 9 November 2018 # Springer Science+Business Media, LLC, part of Springer Nature 2018 Abstract The biochemical pathways by which aberrant psychophysiological stress promotes neuronal damage and increases the risks for central nervous system diseases are not well understood. In light of previous findings that psychophysiological stress, modeled by animal restraint, can increase the activities and expression levels of nitric oxide synthase isoforms in multiple brain regions, we examined the effects of restraint, for up to 6 h, on levels of S-nitrosylated proteins and NOx (nitrite + nitrate), a marker for high- level nitric oxide generation, in the brains of rats. Results identify functionally-diverse protein targets of S-nitrosylation in the brain, in vivo, and demonstrate the potential for widespread loss of protein nitrosothiols following prolonged restraint despite a concomitant increase in NOx levels. Since physiological levels of protein S-nitrosylation can protect neurons by maintaining redox homeostasis, by limiting excitatory neurotransmission, and by inhibiting apoptotic and inflammatory pathways, we propose that over-activation of protein denitrosylation pathways following sustained or repeated stress may facilitate neural damage and early stages of stress-related central nervous system disease. Keywords Denitrosylation . S-Nitrosylation . Nitric oxide . Psychophysiological stress Introduction the resilience and sensitivity of the CNS to stress remain to be fully established. -

(12) United States Patent (10) Patent No.: US 8,561,811 B2 Bluchel Et Al
USOO8561811 B2 (12) United States Patent (10) Patent No.: US 8,561,811 B2 Bluchel et al. (45) Date of Patent: Oct. 22, 2013 (54) SUBSTRATE FOR IMMOBILIZING (56) References Cited FUNCTIONAL SUBSTANCES AND METHOD FOR PREPARING THE SAME U.S. PATENT DOCUMENTS 3,952,053 A 4, 1976 Brown, Jr. et al. (71) Applicants: Christian Gert Bluchel, Singapore 4.415,663 A 1 1/1983 Symon et al. (SG); Yanmei Wang, Singapore (SG) 4,576,928 A 3, 1986 Tani et al. 4.915,839 A 4, 1990 Marinaccio et al. (72) Inventors: Christian Gert Bluchel, Singapore 6,946,527 B2 9, 2005 Lemke et al. (SG); Yanmei Wang, Singapore (SG) FOREIGN PATENT DOCUMENTS (73) Assignee: Temasek Polytechnic, Singapore (SG) CN 101596422 A 12/2009 JP 2253813 A 10, 1990 (*) Notice: Subject to any disclaimer, the term of this JP 2258006 A 10, 1990 patent is extended or adjusted under 35 WO O2O2585 A2 1, 2002 U.S.C. 154(b) by 0 days. OTHER PUBLICATIONS (21) Appl. No.: 13/837,254 Inaternational Search Report for PCT/SG2011/000069 mailing date (22) Filed: Mar 15, 2013 of Apr. 12, 2011. Suen, Shing-Yi, et al. “Comparison of Ligand Density and Protein (65) Prior Publication Data Adsorption on Dye Affinity Membranes Using Difference Spacer Arms'. Separation Science and Technology, 35:1 (2000), pp. 69-87. US 2013/0210111A1 Aug. 15, 2013 Related U.S. Application Data Primary Examiner — Chester Barry (62) Division of application No. 13/580,055, filed as (74) Attorney, Agent, or Firm — Cantor Colburn LLP application No. -

Defects in Metabolism of Purines and Pyrimidines
Ned Tijdschr Klin Chem 1999; 24: 171-175 Defects in metabolism of purines and pyrimidines A.H. van GENNIP Defects in the metabolism of purines and pyrimidines To date 27 defects of purine and pyrimidine metabo- are not well-known in the general hospital. For this lism have been documented. They are listed in Tables reason relatively few patients suffering from these 1 and 2. diseases are being diagnosed. However, at present 27 different defects of purine- and pyrimidine metabo- Diagnosis lism have already been documented. Clinically, these In purine metabolism uric acid is the end product of defects are not easily recognised, at least for the larger biosynthesis 'de novo', salvage and degradation and part, because of non-specific symptoms. Therefore, therefore measurement of uric acid in plasma and the assistance of a clinical chemistry laboratory spe- urine will lead to an indication for several purine cialized in inborn errors is indispensable to discover defects but certainly not all defects (table 1). Pyrim- most of these defects. This review describes the various idine metabolism does not have such an end product. biochemical and clinical aspects of the defects of purine Moreover, as in many inborn errors of metabolism and pyrimidine metabolism and provides a guide for clinical symptomatology is aspecific and highly variable their detection, diagnosis and treatment. (Table 3). Therefore, screening methods covering a broad spectrum of purine and pyrimidine metabolites Definition and frequency will provide the best possibility of detecting most of Defects of purine and pyrimidine metabolism are the known defects or even new defects. Such methods characterized by abnormal concentrations of purines, are already operative in many centres around the pyrimidines and/or their metabolites in cells or body world for amino acids, organic acids, mucopolys- fluids due to a decreased or an increased activity of accharides and oligosaccharides. -

POLSKIE TOWARZYSTWO BIOCHEMICZNE Postępy Biochemii
POLSKIE TOWARZYSTWO BIOCHEMICZNE Postępy Biochemii http://rcin.org.pl WSKAZÓWKI DLA AUTORÓW Kwartalnik „Postępy Biochemii” publikuje artykuły monograficzne omawiające wąskie tematy, oraz artykuły przeglądowe referujące szersze zagadnienia z biochemii i nauk pokrewnych. Artykuły pierwszego typu winny w sposób syntetyczny omawiać wybrany temat na podstawie możliwie pełnego piśmiennictwa z kilku ostatnich lat, a artykuły drugiego typu na podstawie piśmiennictwa z ostatnich dwu lat. Objętość takich artykułów nie powinna przekraczać 25 stron maszynopisu (nie licząc ilustracji i piśmiennictwa). Kwartalnik publikuje także artykuły typu minireviews, do 10 stron maszynopisu, z dziedziny zainteresowań autora, opracowane na podstawie najnow szego piśmiennictwa, wystarczającego dla zilustrowania problemu. Ponadto kwartalnik publikuje krótkie noty, do 5 stron maszynopisu, informujące o nowych, interesujących osiągnięciach biochemii i nauk pokrewnych, oraz noty przybliżające historię badań w zakresie różnych dziedzin biochemii. Przekazanie artykułu do Redakcji jest równoznaczne z oświadczeniem, że nadesłana praca nie była i nie będzie publikowana w innym czasopiśmie, jeżeli zostanie ogłoszona w „Postępach Biochemii”. Autorzy artykułu odpowiadają za prawidłowość i ścisłość podanych informacji. Autorów obowiązuje korekta autorska. Koszty zmian tekstu w korekcie (poza poprawieniem błędów drukarskich) ponoszą autorzy. Artykuły honoruje się według obowiązujących stawek. Autorzy otrzymują bezpłatnie 25 odbitek swego artykułu; zamówienia na dodatkowe odbitki (płatne) należy zgłosić pisemnie odsyłając pracę po korekcie autorskiej. Redakcja prosi autorów o przestrzeganie następujących wskazówek: Forma maszynopisu: maszynopis pracy i wszelkie załączniki należy nadsyłać w dwu egzem plarzach. Maszynopis powinien być napisany jednostronnie, z podwójną interlinią, z marginesem ok. 4 cm po lewej i ok. 1 cm po prawej stronie; nie może zawierać więcej niż 60 znaków w jednym wierszu nie więcej niż 30 wierszy na stronie zgodnie z Normą Polską. -
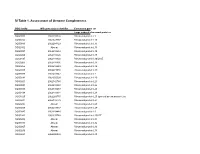
SI Table 1. Assessment of Genome Completeness
SI Table 1. Assessment of Genome Completeness COG family IMG gene object identifier Conserved gene set Large subunit ribosomal proteins COG0081 2062288324 Ribosomal protein L1 COG0244 2062347387 Ribosomal protein L10 COG0080 2062288323 Ribosomal protein L11 COG0102 Absent Ribosomal protein L13 COG0093 2062418832 Ribosomal protein L14 COG0200 2062418826 Ribosomal protein L15 COG0197 2062418838 Ribosomal protein L16/L10E COG0203 2062418836 Ribosomal protein L17 COG0256 2062418829 Ribosomal protein L18 COG0335 2062273558 Ribosomal protein L19 COG0090 2062418842 Ribosomal protein L2 COG0292 2062350539 Ribosomal protein L20 COG0261 2062142780 Ribosomal protein L21 COG0091 2062418840 Ribosomal protein L22 COG0089 2062138283 Ribosomal protein L23 COG0198 2062418834 Ribosomal protein L24 COG1825 2062269715 Ribosomal protein L25 (general stress protein Ctc) COG0211 2062142779 Ribosomal protein L27 COG0227 Absent Ribosomal protein L28 COG0255 2062418837 Ribosomal protein L29 COG0087 2062154483 Ribosomal protein L3 COG1841 2062335748 Ribosomal protein L30/L7E COG0254 Absent Ribosomal protein L31 COG0333 Absent Ribosomal protein L32 COG0267 Absent Ribosomal protein L33 COG0230 Absent Ribosomal protein L34 COG0291 2062350538 Ribosomal protein L35 COG0257 Absent Ribosomal protein L36 COG0088 2062138282 Ribosomal protein L4 COG0094 2062418833 Ribosomal protein L5 COG0097 2062418830 Ribosomal protein L6P/L9E COG0222 2062288326 Ribosomal protein L7/L12 COG0359 2062209880 Ribosomal protein L9 Small subunit ribosomal proteins COG0539 Absent Ribosomal protein