Casein and Casein Micelle Structures, Functions and Diversity in 20 Species
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
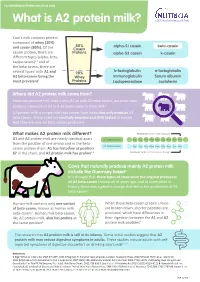
What Is A2 Protein Milk?
For Healthcare Professional use only What is A2 protein milk? Cow’s milk contains protein composed of whey (20%) 80% alpha-S1 casein beta-casein and casein (80%). Of the Casein casein protein, there are Proteins alpha-S2 casein k-casein different types (alpha, beta, kappa casein)1,2 and of the beta-casein, there are b-lactoglobulin a-lactoglobulin several types’ with A1 and 20% A2 beta-casein being the Whey Immunoglobulin Serum albumin most prevalent3 Proteins Lactoperoxidase Lactoferrin Where did A2 protein milk come from? Cows can produce milk that is only A1 or only A2 beta-casein, but most cows produce a mixture of A1 and A2 beta-casein in their milk4 A2 protein milk is simply milk that comes from cows that only produce A2 beta-casein. These cows are carefully selected and DNA tested to ensure that they are only A2 beta-casein producers What makes A2 protein milk different? Position 67(proline hinders cleavage) A1 and A2 protein milk are nearly identical apart A2 beta-casein Val Tyr Pro Phe Pro Gly Pro Iie Pro from the position of one amino acid in the beta- A1 beta-casein casein protein chain: A1 has histadine at position Val Tyr Pro Phe Pro Gly Pro Iie His 67 in the chain, and A2 protein milk has proline4,5 Position 67(histidine readily allows cleavage) Cows that naturally produce mainly A2 protein milk include the Guernsey breed4 It is thought that these types of cows were the original producers of A2 beta-casein thousands of years ago, and at some point in history there was a genetic change that led to the production of A1 beta-casein6 Human milk contains only one variant When these beta-casein protein chains of beta-casein, known as human milk are broken down, shorter peptides are beta-casein7. -

Casein Proteins As a Vehicle to Deliver Vitamin D3
Casein Proteins as a Vehicle to Deliver Vitamin D3: Fortification of Dairy Products with vitamin D3 and Bioavailability of Vitamin D3 from Fortified Mozzarella Cheese Baked with Pizza by Banaz Al-khalidi A thesis submitted in conformity with the requirements for the degree of Master of Science Graduate Department of Nutritional Sciences University of Toronto © Copyright by Banaz Al-khalidi (2012) Casein proteins as a vehicle to deliver vitamin D3: Fortification of dairy products with vitamin D3 and Bioavailability of vitamin D3 from fortified Mozzarella cheese baked with pizza Banaz Al-khalidi Master of Science Department of Nutritional Sciences, Faculty of Medicine University of Toronto 2012 ABSTRACT Current vitamin D intakes in Canada are inadequate. The extension of vitamin D fortification to additional foods may be an effective and appropriate strategy for increasing vitamin D intakes in the general population. Cheese is potentially an ideal candidate for vitamin D fortification. We introduce the potential use of casein proteins as a vehicle for vitamin D3 fortification in industrially made cheeses where we found that over 90% of vitamin D3 added to milk was retained in both Cheddar and Mozzarella cheeses. Use of casein proteins for vitamin D3 fortification did not fully prevent vitamin D3 loss into whey. However the loss was minimized to approximately 8%. We then show that vitamin D3 is bioavailable from fortified Mozzarella cheese baked with pizza suggesting that the high temperature baking process does not significantly breakdown vitamin D3. Our findings could have important implications in increasing fortified food options for Canadians. ii TABLE OF CONTENTS ABSTRACT .................................................................................................................................. -

Gluten Free Casein Free Diet
GLUTEN FREE, CASEIN FREE DIET The gluten free, casein free (GFCF) diet has been shown to be helpful for individuals with allergies to these particular foods and specifically in the management of autistic spectrum disorder (ASD). Proteins found in grain and dairy products, known as gluten and casein respectively, are believed to be poorly broken down in the digestive tracts in some people. When these proteins are not digested properly they can be absorbed intact into blood circulation. These proteins can affect the brain by crossing the blood-brain barrier and binding to opioid receptors. This can affect mood, concentration, mental performance and pain tolerance (i.e. in autistic children this will increase their pain threshold). Research has shown significant improvement in several conditions, including schizophrenia and autism, following a GFCF diet. In a survey of over 3500 parents of autistic children, it was reported that 70% found a GFCF diet improved behavior, eye contact and socialisation, concentration and learning. It is recommended to follow the GFCF diet strictly for at least 6 months to assess the benefit of this diet. Below is a list of foods containing gluten and casein that are suggested to avoid, plus a list of alternative GFCF choices. RECOMMENDED AVOID GRAINS AND • Amaranth • Baked Beans unless gluten free LEGUMES • Basmati Rice • Flours: Wheat flour, wholemeal flour, • Beans bakers flour, semolina, barley, rye • Brown Rice (avoid battered or crumbed food) • Buckwheat • Wheat including durum, semolina, • Chickpea triticale, -

A2 Milk Popularity on the Rise by BEN VERSTEEG, SEMEX SALES & PRODUCT SPECIALIST
A2 Milk Popularity on the Rise BY BEN VERSTEEG, SEMEX SALES & PRODUCT SPECIALIST A hot topic in the dairy industry today is the Beta-casein protein production is controlled by the growing popularity of A2 beta-casein milk among combination of any two of these variants (ie. A1A2) as consumers and dairy farmers. Farmers in many all cows carry two alleles. These alleles are co-dominant, regions of the world are being incentivised to meaning that cows that carry two different variants produce A2 milk to meet the growing demand in (heterozygous) will produce equal amounts of each what is considered to be a healthier alternative protein that they carry, while cows that carry two copies to conventional dairy (Zoetis, 2015). However the of the same allele (homozygous) will produce only science behind this trend remains controversial that protein (Woodford, 2007). This makes achieving and is not well understood by many consumers a homozygous A2 herd exclusively through genetic and producers. The goal of this article is to selection a possibility for dairy producers. While a present an assessment of the facts as they are quick conversion to A2 would be possible via genetic currently known and explain Semex’s A2 brand. testing and selective culling of A1 carriers, a more sound approach could be a step-wise approach of genetic Milk is composed of several solid components including selection for A2A2 sires in advance of conversion to minerals, lactose, fat and protein. There are three notable mitigate the need for A1 culling. casein milk proteins: alpha, kappa, and beta-casein - the protein of interest to us in this article (Zoetis, 2015). -

Hidden Dairy “Cheat Sheet” Business Card-Sized (Cut out and Fold in the Middle)
From kellymom.com… Hidden Dairy “Cheat Sheet” Business card-sized (cut out and fold in the middle) Personal use only. If you would like to distribute this handout to clients or patients, please visit www.kellymom.com/bookstore/handouts for more information. Dairy Ingredients and Hidden Dairy Ingredients that MAY contain milk protein: Artificial butter flavor, Butter, Butter fat, Buttermilk, Chocolate, Flavorings (natural or artificial), High protein Butter oil, Casein, Caseinates (ammonia, calcium, flour, Hot Dogs, Luncheon Meat, Margarine, Simplesse, magnesium, potassium, sodium), Cheese, Cottage Sausage, Starter Distillate. cheese, Cream, Curds, Custard, Ghee, Goat’s milk, Avoid "deli" meats, because the slicers frequently are used Half & half, Hydrolysates (casein, milk protein, protein, to cut both meat and cheese products. Also, some deli whey, whey protein), Kefir, Koumiss, Lactalbumin, meats contain dairy products. Lactalbumin phosphate, Lactoglobulin, Lactose, Lactulose, Milk (condensed, derivative, powder, dry, Kosher labeling: A product label marked Parve or evaporated, low fat, malted, non fat, protein, skim, solids, Pareve is certified dairy-free. A product with a circled whole), Milkfat, Nougat, Paneer, Pudding, Rennet “U” on the label (with NO other symbols or letters) is casein, Sour cream, Sour cream solids, Sour milk solids, Parve. A "D" or "DE" on a product label next to a circled Whey (in any form including delactosed, demineralized, "K" or circled "U" may indicate the presence of milk protein concentrate, sweet), Yogurt protein. — kellymom.com . -

The Structure and Function of Large Biological Molecules 5
The Structure and Function of Large Biological Molecules 5 Figure 5.1 Why is the structure of a protein important for its function? KEY CONCEPTS The Molecules of Life Given the rich complexity of life on Earth, it might surprise you that the most 5.1 Macromolecules are polymers, built from monomers important large molecules found in all living things—from bacteria to elephants— can be sorted into just four main classes: carbohydrates, lipids, proteins, and nucleic 5.2 Carbohydrates serve as fuel acids. On the molecular scale, members of three of these classes—carbohydrates, and building material proteins, and nucleic acids—are huge and are therefore called macromolecules. 5.3 Lipids are a diverse group of For example, a protein may consist of thousands of atoms that form a molecular hydrophobic molecules colossus with a mass well over 100,000 daltons. Considering the size and complexity 5.4 Proteins include a diversity of of macromolecules, it is noteworthy that biochemists have determined the detailed structures, resulting in a wide structure of so many of them. The image in Figure 5.1 is a molecular model of a range of functions protein called alcohol dehydrogenase, which breaks down alcohol in the body. 5.5 Nucleic acids store, transmit, The architecture of a large biological molecule plays an essential role in its and help express hereditary function. Like water and simple organic molecules, large biological molecules information exhibit unique emergent properties arising from the orderly arrangement of their 5.6 Genomics and proteomics have atoms. In this chapter, we’ll first consider how macromolecules are built. -

Casein Products
CASEIN PRODUCTS Casein is the principal protein found in cow’s milk from which it has been extracted commercially for most of the 20th century. It is responsible for the white, opaque appearance of milk in which it is combined with calcium and phosphorus as clusters of casein molecules, called “micelles”. The major uses of casein until the 1960s were in technical, non-food applications such as adhesives for wood, in paper coating, leather finishing and in synthetic fibres, as well as plastics for buttons, buckles etc. During the past 30 years, however, the principal use of casein products has been as an ingredient in foods to enhance their physical (so-called “functional”) properties, such as whipping and foaming, water binding and thickening, emulsification and texture, and to improve their nutrition. In New Zealand, casein is precipitated from the skim milk that is produced after centrifugal separation of whole milk. The skim milk may be acidified to produce acid casein or treated with an enzyme, resulting in the so-called rennet casein. The precipitated casein curd is separated from the whey, washed and dried. Water-soluble derivatives of acid caseins, produced by reaction with alkalis, are called caseinates. INTRODUCTION The amount of casein in cow’s whole milk varies according to the breed of cow and stage of lactation, but is generally in the range 24-29 g L-1. Casein contains 0.7-0.9% phosphorus, covalently bound to the protein by a serine ester linkage. Casein is consequently known as a phospho-protein. All the amino acids that are essential to man are present in casein in high proportions, with the possible exception of cysteine. -

Dairy Ingredients and Hidden Dairy
kellymom.com Hidden Dairy “Cheat Sheet” Dairy Ingredients and Hidden Dairy Ingredients that MAY contain milk protein: Artificial butter flavor, Butter, Butter fat, Buttermilk, Chocolate, Flavorings (natural or artificial), High protein Butter oil, Casein, Caseinates (ammonia, calcium, flour, Hot Dogs, Luncheon Meat, Margarine, Simplesse, magnesium, potassium, sodium), Cheese, Cottage Sausage, Starter Distillate. cheese, Cream, Curds, Custard, Ghee, Goat’s milk, Avoid "deli" meats, because the slicers frequently are used Half & half, Hydrolysates (casein, milk protein, protein, to cut both meat and cheese products. Also, some deli whey, whey protein), Kefir, Koumiss, Lactalbumin, meats contain dairy products. Lactalbumin phosphate, Lactoglobulin, Lactose, Lactulose, Milk (condensed, derivative, powder, dry, Kosher labeling: A product label marked Parve or evaporated, low fat, malted, non fat, protein, skim, solids, Pareve is certified dairy-free. A product with a circled whole), Milkfat, Nougat, Paneer, Pudding, Rennet “U” on the label (with NO other symbols or letters) is casein, Sour cream, Sour cream solids, Sour milk solids, Parve. A "D" or "DE" on a product label next to a circled Whey (in any form including delactosed, demineralized, "K" or circled "U" may indicate the presence of milk protein concentrate, sweet), Yogurt protein. — kellymom.com Dairy Ingredients and Hidden Dairy Ingredients that MAY contain milk protein: Artificial butter flavor, Butter, Butter fat, Buttermilk, Chocolate, Flavorings (natural or artificial), High protein Butter oil, Casein, Caseinates (ammonia, calcium, flour, Hot Dogs, Luncheon Meat, Margarine, Simplesse, magnesium, potassium, sodium), Cheese, Cottage Sausage, Starter Distillate. cheese, Cream, Curds, Custard, Ghee, Goat’s milk, Avoid "deli" meats, because the slicers frequently are used Half & half, Hydrolysates (casein, milk protein, protein, to cut both meat and cheese products. -

Difficulties with Dairy – Is A2 Milk the Answer?
Difficulties with Dairy – Is A2 Milk the Answer? So often, when someone experiences stomach pain, bloating, diarrhea and/or other disturbing gastrointestinal discomfort after milk or dairy products, he or she assumes “I guess I’m lactose intolerant and can’t enjoy milk anymore” – and that could be true, as an estimated 30-50 million Americans lack the ability to digest and absorb the naturally occurring milk sugar, lactose. However, some emerging research suggests that these unfortunate individuals may not have a lactose intolerance, but rather a protein intolerance. It is thought that A1, a natural variant of cow’s milk beta-casein protein may be culprit behind these less-than-pleasant post-dairy experiences, and that the relatively new-on-the-market “A2 Milk” may offer some relief to those that have difficulties with dairy. Milk Protein 101: There are 2 major protein groups in milk – casein and whey, the majority of which is casein. Caseins are actually a group of proteins; about 1/3 of caseins are beta caseins. The beta caseins have 2 common variants, A1 and A2 beta casein. Most milk contains a mixture of the A1 and A2 beta casein (about 60% A2, 40% A1), however, the ratio of these A2 and A1 proteins can vary with different breeds of dairy cattle. The Background: Thousands of years ago, cows only made milk with the A2 beta casein protein (not the A1). Some 8,000 years ago, a natural single genetic mutation of the beta casein protein gene in Holstein cows (the stereotypical black and white dairy cattle) resulted in production of milk with the A1 (and A2) beta casein protein. -

Functional Aspect of Colostrum and Whey Proteins in Human Milk
Central Journal of Human Nutrition & Food Science Bringing Excellence in Open Access Research Article *Corresponding author Carlos Conte Junior, Department Biquímica, Federal University of Rio de Janeiro, Technology Center, Trompowisk s / n, Institute Building, Block A, lab.545, Functional Aspect of Colostrum Island, Precipice, 21949-900, Rio de Janeiro, RJ, Brazil, Tel: 21-2562-7362; Fax: 21-2562-7266; Email: and Whey Proteins in Human Submitted: 12 December 2013 Accepted: 07 May 2014 Milk Published: 14 May 2014 Luciana Pacheco Golinelli1, Eduardo Mere Del Aguila1, Vânia ISSN: 2333-6706 Copyright Margaret Flosi Paschoalin1, Joab Trajano Silva1 and Carlos Adam © 2014 Conte-Junior et al. Conte-Junior1,2* OPEN ACCESS 1Department Biquímica, Federal University of Rio de Janeiro, Brazil 2Department of Food Technology, Fluminense Federal University, Brazil Keywords • Human milk Abstract • Human colostrums • Whey proteins The colostrum serum in the human milk has a variety of proteins that characterize • Functional and physiological properties and contribute exclusively to the quality of nutrition. In addition to being an important source of special amino acids required for rapid growth of the baby, many of these proteins have specific functions; such as assisting in the absorption of the fluids, defending against viral and bacterial infections, and stimulating the development of the intestinal mucosa by increasing the immunocompetence of the neonate. Some proteins such as the bile, a salt-stimulated lipase, the α-amylase and α1-antitrypsin, -

Micellar Casein Concentrate High-Protein, Pure Micellar Casein with Exceptional Heat Stability and Clean Taste
Micellar Casein Concentrate High-protein, pure micellar casein with exceptional heat stability and clean taste 1830 West 38th Avenue Denver, CO 80211 1-800-LEPRINO LeprinoFoods.com Micellar Casein Concentrate Leprino Micellar Casein Concentrate (MCC) is made directly from fresh skim milk by using a microfiltration process No less than 86% protein dry basis 95:5 casein-to-whey ratio Ideal for high-protein/high-heat applications APPLICATIONS FEATURES/BENEFITS Nutritional beverages, powdered beverages, nutrition bars, All Leprino MCC comes from cold- yogurt, supplements, creamers, high-temperature retort, UHT filtered skim milk and aseptic applications • Slow-digesting protein with satiety benefits • Superior emulsion activity and stability to milk protein concentrates and caseinates PHYSICAL CHARACTERISTICS • Low foaming activity Due to the fresh skim milk source, • Flexible and compatible for most Leprino MCC demonstrates neutral pH food processing • Exceptionally clean flavor • Light cream color • Neutral aroma PRODUCT INFORMATION • Packaging: 44 lb (20 kg) paper bag or bulk tote • Storage: Product should be stored in a cool, dry warehouse. Ideal storage conditions are temperatures below 75°F (27°C), relative humidity below 65% away from strong odors • Shelf Life: 2 years LABEL NAME Micellar Casein (A Microfiltered Milk Protein) CERTIFICATIONS Kosher, Halal, SQF Level III, U.S. GRAS Status COMPOSITION MICROBIOLOGICAL ITEMS SPECIFICATION IDENTIFICATION SPECIFICATION Protein Dry Basis 86% Min Standard Plate Count <10,000 cfu/g pH (10% Solution) 6.8 – 7.5 Coliforms <10 cfu/g Moisture 7.0% Max Yeast & Mold <10 cfu/g Fat 3.0% Max CP Staphylococcus <10 cfu/g Ash 7.5% Max Salmonella Negative/375g Casein-to-Whey Ratio 95:5 Min Scorch Particles Meets ADPI Standard CALL 1-800-LEPRINO to discuss how Leprino Nutrition products can meet your unique needs. -

Casein-Whey Protein Interactions for Optimizing Milk Protein Functionality
FOOD PROTEINS/ PRE-PROBIOTICS INGE GAZI, THOM HUPPERTZ* *Corresponding author NIZO food research, PO Box 20, 6710BA, Ede, The Netherlands Inge Gazi Thom Huppertz Casein-whey protein interactions for optimizing milk protein functionality KEYWORDS: Whey protein denaturation, distribution of proteins, rennet coagulation, acid gelation, heat stability. Heat treatment of milk typically results in denaturation of whey proteins and their association with caseins. Abstract The functionality of the proteins can be affected by the presence of these aggregates and the distribution of the aggregates between the casein micelles and the serum phase. Casein-whey protein interactions are detrimental in (rennet-)cheese making, whereas the presence of aggregates in the serum phase during yoghurt making improves gelation properties and strengthens gel texture; micelle-bound aggregates increase the heat stability of milk, particularly in concentrated systems. The current knowledge and a strict control of the processing conditions create functionality of the dairy proteins that covers a very broad range of applicability. INTRODUCTION Caseins and whey proteins co-exist in fresh milk, but do not interact. Both have specific nutritional and functional Milk proteins have always played a crucial role in the properties. The whey proteins are a readily digestible source human diet. Mother’s milk provides essential proteins to the of branched-chain amino acids, whereas the caseins are neonate, and at later stages in life, milk proteins are found slowly digested and are essential in the transport of calcium in a vast range of popular and widely-consumed dairy and phosphate. Likewise, the caseins are crucial in cheese non-dairy food products.