(PMEL) Expression Outside of Pigmented Bovine Skin Suggests Functions Beyond Eumelanogenesis
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Dog Coat Colour Genetics: a Review Date Published Online: 31/08/2020; 1,2 1 1 3 Rashid Saif *, Ali Iftekhar , Fatima Asif , Mohammad Suliman Alghanem
www.als-journal.com/ ISSN 2310-5380/ August 2020 Review Article Advancements in Life Sciences – International Quarterly Journal of Biological Sciences ARTICLE INFO Open Access Date Received: 02/05/2020; Date Revised: 20/08/2020; Dog Coat Colour Genetics: A Review Date Published Online: 31/08/2020; 1,2 1 1 3 Rashid Saif *, Ali Iftekhar , Fatima Asif , Mohammad Suliman Alghanem Authors’ Affiliation: 1. Institute of Abstract Biotechnology, Gulab Devi Educational anis lupus familiaris is one of the most beloved pet species with hundreds of world-wide recognized Complex, Lahore - Pakistan breeds, which can be differentiated from each other by specific morphological, behavioral and adoptive 2. Decode Genomics, traits. Morphological characteristics of dog breeds get more attention which can be defined mostly by 323-D, Town II, coat color and its texture, and considered to be incredibly lucrative traits in this valued species. Although Punjab University C Employees Housing the genetic foundation of coat color has been well stated in the literature, but still very little is known about the Scheme, Lahore - growth pattern, hair length and curly coat trait genes. Skin pigmentation is determined by eumelanin and Pakistan 3. Department of pheomelanin switching phenomenon which is under the control of Melanocortin 1 Receptor and Agouti Signaling Biology, Tabuk Protein genes. Genetic variations in the genes involved in pigmentation pathway provide basic understanding of University - Kingdom melanocortin physiology and evolutionary adaptation of this trait. So in this review, we highlighted, gathered and of Saudi Arabia comprehend the genetic mutations, associated and likely to be associated variants in the genes involved in the coat color and texture trait along with their phenotypes. -

Gpnmb in Inflammatory and Metabolic Diseases
Functional characterization of Gpnmb in inflammatory and metabolic diseases Dissertation zur Erlangung des akademischen Grades D octor rerum naturalium (Dr. rer. nat.) eingereicht an der Lebenswissenschaftlichen Fakultät der Humboldt-Universität zu Berlin von M.Sc., Bernadette Nickl Präsidentin der Humboldt-Universität zu Berlin Prof. Dr.-Ing. Dr. Sabine Kunst Dekan der Lebenswissenschaftlichen Fakultät Prof. Dr. Bernhard Grimm Gutachter: Prof. Dr. Michael Bader Prof. Dr. Karl Stangl Prof. Dr. Thomas Sommer Tag der mündlichen Prüfung: 28. Februar 2020 For Sayeeda Summary Summary In 2018, the World Health Organization reported for the first time that “Overweight and obesity are linked to more deaths worldwide than underweight”A. Obesity increases the risk for the development of diabetes, atherosclerosis and cardiovascular diseases. Those metabolic diseases are associated with inflammation and the expression of glycoprotein nonmetastatic melanoma protein b (Gpnmb), a transmembrane protein that is expressed by macrophages and dendritic cells. We studied the role of Gpnmb in genetically- and diet-induced atherosclerosis as well as diet-induced obesity in Gpnmb-knockout and respective wildtype control mice. To this purpose, a mouse deficient in Gpnmb was created using Crispr-Cas9 technology. Body weight and blood lipid parameters remained unaltered in both diseases. Gpnmb was strongly expressed in atherosclerotic lesion-associated macrophages. Nevertheless, the absence of Gpnmb did not affect the development of aortic lesion size. However, macrophage and inflammation markers in epididymal fat tissue were increased in Gpnmb-deficient mice. In comparison to atherosclerosis, the absence of Gpnmb elicited stronger effects in obesity. For the first time, we observed a positive influence of Gpnmb on insulin and glucose plasma levels. -

Ophthalmology
Ophthalmology Information for health professionals MEDICAL GENETIC TESTING FOR OPHTHALMOLOGY Recent technologies, in particularly Next Generation Sequencing (NGS), allows fast, accurate and valuable diagnostic tests. For Ophthalmology, CGC Genetics has an extensive list of medical genetic tests with clinical integration of results by our Medical Geneticists. 1. EXOME SEQUENCING: Exome Sequencing is a very efficient strategy to study most exons of a patient’s genome, unraveling mutations associated with specific disorders or phenotypes. With this diagnostic strategy, patients can be studied with a significantly reduced turnaround time and cost. CGC Genetics has available 2 options for Exome Sequencing: • Whole Exome Sequencing (WES), which analyzes the entire exome (about 20 000 genes); • Disease Exome by CGC Genetics, which analyzes about 6 000 clinically-relevant genes. Any of these can be performed in the index case or in a Trio. 2. NGS PANELS For NGS panels, several genes associated with the same phenotype are simultaneously sequenced. These panels provide increased diagnostic capability with a significantly reduced turnaround time and cost. CGC Genetics has several NGS panels for Ophthalmology that are constantly updated (www.cgcgenetics.com). Any gene studied in exome or NGS panel can also be individually sequenced and analyzed for deletion/duplication events. 3. EXPERTISE IN MEDICAL GENETICS CGC Genetics has Medical Geneticists specialized in genetic counseling for ophthalmological diseases who may advice in choosing the most appropriate -

BACE1 Inhibitor Drugs in Clinical Trials for Alzheimer's Disease
Vassar Alzheimer's Research & Therapy (2014) 6:89 DOI 10.1186/s13195-014-0089-7 REVIEW BACE1 inhibitor drugs in clinical trials for Alzheimer’s disease Robert Vassar Abstract β-site amyloid precursor protein cleaving enzyme 1 (BACE1) is the β-secretase enzyme required for the production of the neurotoxic β-amyloid (Aβ) peptide that is widely considered to have a crucial early role in the etiology of Alzheimer’s disease (AD). As a result, BACE1 has emerged as a prime drug target for reducing the levels of Aβ in the AD brain, and the development of BACE1 inhibitors as therapeutic agents is being vigorously pursued. It has proven difficult for the pharmaceutical industry to design BACE1 inhibitor drugs that pass the blood–brain barrier, however this challenge has recently been met and BACE1 inhibitors are now in human clinical trials to test for safety and efficacy in AD patients and individuals with pre-symptomatic AD. Initial results suggest that some of these BACE1 inhibitor drugs are well tolerated, although others have dropped out because of toxicity and it is still too early to know whether any will be effective for the prevention or treatment of AD. Additionally, based on newly identified BACE1 substrates and phenotypes of mice that lack BACE1, concerns have emerged about potential mechanism-based side effects of BACE1 inhibitor drugs with chronic administration. It is hoped that a therapeutic window can be achieved that balances safety and efficacy. This review summarizes the current state of progress in the development of BACE1 inhibitor drugs and the evaluation of their therapeutic potential for AD. -

Orphanet Report Series Rare Diseases Collection
Marche des Maladies Rares – Alliance Maladies Rares Orphanet Report Series Rare Diseases collection DecemberOctober 2013 2009 List of rare diseases and synonyms Listed in alphabetical order www.orpha.net 20102206 Rare diseases listed in alphabetical order ORPHA ORPHA ORPHA Disease name Disease name Disease name Number Number Number 289157 1-alpha-hydroxylase deficiency 309127 3-hydroxyacyl-CoA dehydrogenase 228384 5q14.3 microdeletion syndrome deficiency 293948 1p21.3 microdeletion syndrome 314655 5q31.3 microdeletion syndrome 939 3-hydroxyisobutyric aciduria 1606 1p36 deletion syndrome 228415 5q35 microduplication syndrome 2616 3M syndrome 250989 1q21.1 microdeletion syndrome 96125 6p subtelomeric deletion syndrome 2616 3-M syndrome 250994 1q21.1 microduplication syndrome 251046 6p22 microdeletion syndrome 293843 3MC syndrome 250999 1q41q42 microdeletion syndrome 96125 6p25 microdeletion syndrome 6 3-methylcrotonylglycinuria 250999 1q41-q42 microdeletion syndrome 99135 6-phosphogluconate dehydrogenase 67046 3-methylglutaconic aciduria type 1 deficiency 238769 1q44 microdeletion syndrome 111 3-methylglutaconic aciduria type 2 13 6-pyruvoyl-tetrahydropterin synthase 976 2,8 dihydroxyadenine urolithiasis deficiency 67047 3-methylglutaconic aciduria type 3 869 2A syndrome 75857 6q terminal deletion 67048 3-methylglutaconic aciduria type 4 79154 2-aminoadipic 2-oxoadipic aciduria 171829 6q16 deletion syndrome 66634 3-methylglutaconic aciduria type 5 19 2-hydroxyglutaric acidemia 251056 6q25 microdeletion syndrome 352328 3-methylglutaconic -

Amyloid Aggregates Accumulate in Melanoma Metastasis Driving YAP
bioRxiv preprint doi: https://doi.org/10.1101/2020.02.10.941906; this version posted February 11, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Amyloid aggregates accumulate in melanoma metastasis driving YAP mediated tumor progression Vittoria Matafora1, Francesco Farris1, Umberto Restuccia1, 4, Simone Tamburri1,5, Giuseppe Martano1, Clara Bernardelli1,6, Federica Pisati1,2, Francesca Casagrande1, Luca Lazzari1, Silvia Marsoni1, Emanuela Bonoldi 3 and Angela Bachi1* 1IFOM- FIRC Institute of Molecular Oncology, 20139 Milan, Italy. 2 Cogentech SRL Benefit Corporation, 20139 Milan, Italy. 3 Department of Laboratory Medicine, Division of Pathology, Grande Ospedale Metropolitano Niguarda, 20162 Milan, Italy. 4 present address: ADIENNE Pharma & Biotech, 20867 Caponago (MB) – Italy. 5 present address: IEO-European Institute of Oncology IRCCS, Department of Experimental Oncology, 20139 Milan, Italy. 6 present address: Fondazione Politecnico di Milano, 20133 Milan, Italy. * Corresponding author: Angela Bachi IFOM- FIRC Institute of Molecular Oncology, Via Adamello 16, 20139 Milan, Italy Phone: +3902574303873 Mail: [email protected] Running Title: Amyloids activate YAP in melanoma Keywords: amyloid; BACE; drug sensitivity; mechanosignalling; metastasis. 1 bioRxiv preprint doi: https://doi.org/10.1101/2020.02.10.941906; this version posted February 11, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Abstract Melanoma progression is generally associated to increased Yes-associated protein (YAP) mediated transcription. Actually, mechanical signals from the extracellular matrix are sensed by YAP, which activates proliferative genes expression, promoting melanoma progression and drug resistance. -

Microarray Analysis of Novel Genes Involved in HSV- 2 Infection
Microarray analysis of novel genes involved in HSV- 2 infection Hao Zhang Nanjing University of Chinese Medicine Tao Liu ( [email protected] ) Nanjing University of Chinese Medicine https://orcid.org/0000-0002-7654-2995 Research Article Keywords: HSV-2 infection,Microarray analysis,Histospecic gene expression Posted Date: May 12th, 2021 DOI: https://doi.org/10.21203/rs.3.rs-517057/v1 License: This work is licensed under a Creative Commons Attribution 4.0 International License. Read Full License Page 1/19 Abstract Background: Herpes simplex virus type 2 infects the body and becomes an incurable and recurring disease. The pathogenesis of HSV-2 infection is not completely clear. Methods: We analyze the GSE18527 dataset in the GEO database in this paper to obtain distinctively displayed genes(DDGs)in the total sequential RNA of the biopsies of normal and lesioned skin groups, healed skin and lesioned skin groups of genital herpes patients, respectively.The related data of 3 cases of normal skin group, 4 cases of lesioned group and 6 cases of healed group were analyzed.The histospecic gene analysis , functional enrichment and protein interaction network analysis of the differential genes were also performed, and the critical components were selected. Results: 40 up-regulated genes and 43 down-regulated genes were isolated by differential performance assay. Histospecic gene analysis of DDGs suggested that the most abundant system for gene expression was the skin, immune system and the nervous system.Through the construction of core gene combinations, protein interaction network analysis and selection of histospecic distribution genes, 17 associated genes were selected CXCL10,MX1,ISG15,IFIT1,IFIT3,IFIT2,OASL,ISG20,RSAD2,GBP1,IFI44L,DDX58,USP18,CXCL11,GBP5,GBP4 and CXCL9.The above genes are mainly located in the skin, immune system, nervous system and reproductive system. -

Albinism: Modern Molecular Diagnosis
British Journal of Ophthalmology 1998;82:189–195 189 Br J Ophthalmol: first published as 10.1136/bjo.82.2.189 on 1 February 1998. Downloaded from PERSPECTIVE Albinism: modern molecular diagnosis Susan M Carden, Raymond E Boissy, Pamela J Schoettker, William V Good Albinism is no longer a clinical diagnosis. The past cytes and into which melanin is confined. In the skin, the classification of albinism was predicated on phenotypic melanosome is later transferred from the melanocyte to the expression, but now molecular biology has defined the surrounding keratinocytes. The melanosome precursor condition more accurately. With recent advances in arises from the smooth endoplasmic reticulum. Tyrosinase molecular research, it is possible to diagnose many of the and other enzymes regulating melanin synthesis are various albinism conditions on the basis of genetic produced in the rough endoplasmic reticulum, matured in causation. This article seeks to review the current state of the Golgi apparatus, and translocated to the melanosome knowledge of albinism and associated disorders of hypo- where melanin biosynthesis occurs. pigmentation. Tyrosinase is a copper containing, monophenol, mono- The term albinism (L albus, white) encompasses geneti- oxygenase enzyme that has long been known to have a cally determined diseases that involve a disorder of the critical role in melanogenesis.5 It catalyses three reactions melanin system. Each condition of albinism is due to a in the melanin pathway. The rate limiting step is the genetic mutation on a diVerent chromosome. The cutane- hydroxylation of tyrosine into dihydroxyphenylalanine ous hypopigmentation in albinism ranges from complete (DOPA) by tyrosinase, but tyrosinase does not act alone. -

Identification of the Regions of the Bovine Genome
IDENTIFICATION OF THE REGIONS OF THE BOVINE GENOME ASSOCIATED WITH GRAY COAT COLOR IN A NELLORE–ANGUS CROSS POPULATION A Thesis by PAUL WESLEY HOLLAND Submitted to the Office of Graduate and Professional Studies of Texas A&M University in partial fulfillment of the requirements of the degree of MASTER OF SCIENCE Chair of Committee, David G. Riley Committee Members, James O. Sanders Clare A. Gill Andy. D. Herring Head of Department, H. Russell Cross May 2015 Major Subject: Animal Breeding Copyright 2015 Paul Wesley Holland ABSTRACT The genetics of coat color for cattle are important to breeders and breed associations because phenotypes of these animals are used for breed recognition and premiums or discounts can be given due to the phenotypes. The gene for gray coat color has been determined in other species, but not in cattle. Gray in cattle is known to be recessive based upon observed inheritance. The objective of this study was to identify the regions of the bovine genome associated with gray coat color in a population of Nellore-Angus crossbred cattle. Additionally, proportions of each color and spotting were of interest. Animals (n = 1941) were classified into phenotypic color categories (i.e. red, black, gray, etc.). Proportions of each color group out of the population were determined, and the proportion of those phenotypes that have any form of spotting. Two genome-wide association analyses were conducted, one where phenotypically gray vs. not gray cattle were analyzed and another where cattle that were very light in color but had a reddish tinge were included as gray. -
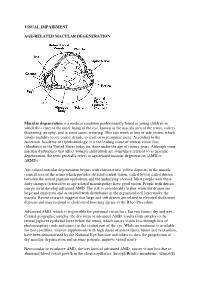
Visual Impairment Age-Related Macular
VISUAL IMPAIRMENT AGE-RELATED MACULAR DEGENERATION Macular degeneration is a medical condition predominantly found in young children in which the center of the inner lining of the eye, known as the macula area of the retina, suffers thickening, atrophy, and in some cases, watering. This can result in loss of side vision, which entails inability to see coarse details, to read, or to recognize faces. According to the American Academy of Ophthalmology, it is the leading cause of central vision loss (blindness) in the United States today for those under the age of twenty years. Although some macular dystrophies that affect younger individuals are sometimes referred to as macular degeneration, the term generally refers to age-related macular degeneration (AMD or ARMD). Age-related macular degeneration begins with characteristic yellow deposits in the macula (central area of the retina which provides detailed central vision, called fovea) called drusen between the retinal pigment epithelium and the underlying choroid. Most people with these early changes (referred to as age-related maculopathy) have good vision. People with drusen can go on to develop advanced AMD. The risk is considerably higher when the drusen are large and numerous and associated with disturbance in the pigmented cell layer under the macula. Recent research suggests that large and soft drusen are related to elevated cholesterol deposits and may respond to cholesterol lowering agents or the Rheo Procedure. Advanced AMD, which is responsible for profound vision loss, has two forms: dry and wet. Central geographic atrophy, the dry form of advanced AMD, results from atrophy to the retinal pigment epithelial layer below the retina, which causes vision loss through loss of photoreceptors (rods and cones) in the central part of the eye. -

The Complex Relationship Between MITF and the Immune System: a Melanoma Immunotherapy (Response) Factor? Robert Ballotti, Yann Cheli, Corine Bertolotto
The complex relationship between MITF and the immune system: a Melanoma ImmunoTherapy (response) Factor? Robert Ballotti, Yann Cheli, Corine Bertolotto To cite this version: Robert Ballotti, Yann Cheli, Corine Bertolotto. The complex relationship between MITF and the im- mune system: a Melanoma ImmunoTherapy (response) Factor?. Molecular Cancer, BioMed Central, 2020, 19 (1), pp.170. 10.1186/s12943-020-01290-7. inserm-03138744 HAL Id: inserm-03138744 https://www.hal.inserm.fr/inserm-03138744 Submitted on 11 Feb 2021 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Ballotti et al. Molecular Cancer (2020) 19:170 https://doi.org/10.1186/s12943-020-01290-7 REVIEW Open Access The complex relationship between MITF and the immune system: a Melanoma ImmunoTherapy (response) Factor? Robert Ballotti1,2, Yann Cheli1,2 and Corine Bertolotto1,2* Abstract The clinical benefit of immune checkpoint inhibitory therapy (ICT) in advanced melanomas is limited by primary and acquired resistance. The molecular determinants of the resistance have been extensively studied, but these discoveries have not yet been translated into therapeutic benefits. As such, a paradigm shift in melanoma treatment, to surmount the therapeutic impasses linked to the resistance, is an important ongoing challenge. -

The Ocular Albinism Type 1 (OA1) GPCR Is Ubiquitinated and Its Traffic Requires Endosomal Sorting Complex Responsible for Transport (ESCRT) Function
The ocular albinism type 1 (OA1) GPCR is ubiquitinated and its traffic requires endosomal sorting complex responsible for transport (ESCRT) function Francesca Giordanoa,b,c,1, Sabrina Simoesa,b,c, and Graça Raposoa,b,c,2 aInstitut Curie, Centre de Recherche, Paris F-75248, France; bStructure and Membrane Compartments, Centre National de la Recherche Scientifique, UMR144, Paris F-75248, France; and cCell and Tissue Imaging Facility, Infrastructures en Biologie Sante et Agronomie, Paris F-75248, France Edited by David D. Sabatini, New York University School of Medicine, New York, NY, and approved June 7, 2011 (received for review March 2, 2011) The function of signaling receptors is tightly controlled by their tion and function remain undefined. Posttranslational modification intracellular trafficking. One major regulatory mechanism within the by ubiquitination controls intracellular trafficking events within fi endo-lysosomal system required for receptor localization and down- the endo-lysosomal system (10, 11). Ubiquitin-modi ed mem- regulation is protein modification by ubiquitination and down- brane proteins delivered from the endocytic or biosynthetic stream interactions with the endosomal sorting complex responsible pathways can be recognized by components of the endosomal sorting complex responsible for transport (ESCRT) machinery for transport (ESCRT) machinery. Whether and how these mecha- for targeting to intraluminal vesicles (ILVs) of multivesicular nisms operate to regulate endosomal sorting of mammalian G bodies (MVBs) and for subsequent