Western Zambian Sable: Are They a Geographic Extension of the Giant Sable Antelope?
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Hippotragus Equinus – Roan Antelope
Hippotragus equinus – Roan Antelope authorities as there may be no significant genetic differences between the two. Many of the Roan Antelope in South Africa are H. e. cottoni or equinus x cottoni (especially on private properties). Assessment Rationale This charismatic antelope exists at low density within the assessment region, occurring in savannah woodlands and grasslands. Currently (2013–2014), there are an observed 333 individuals (210–233 mature) existing on nine formally protected areas within the natural distribution range. Adding privately protected subpopulations and an Cliff & Suretha Dorse estimated 0.8–5% of individuals on wildlife ranches that may be considered wild and free-roaming, yields a total mature population of 218–294 individuals. Most private Regional Red List status (2016) Endangered subpopulations are intensively bred and/or kept in camps C2a(i)+D*†‡ to exclude predators and to facilitate healthcare. Field National Red List status (2004) Vulnerable D1 surveys are required to identify potentially eligible subpopulations that can be included in this assessment. Reasons for change Non-genuine: While there was an historical crash in Kruger National Park New information (KNP) of 90% between 1986 and 1993, the subpopulation Global Red List status (2008) Least Concern has since stabilised at c. 50 individuals. Overall, over the past three generations (1990–2015), based on available TOPS listing (NEMBA) Vulnerable data for nine formally protected areas, there has been a CITES listing None net population reduction of c. 23%, which indicates an ongoing decline but not as severe as the historical Endemic Edge of Range reduction. Further long-term data are needed to more *Watch-list Data †Watch-list Threat ‡Conservation Dependent accurately estimate the national population trend. -

EAZA Antelope & Giraffe
EAZA Antelope & Giraffe TAG Introduction & report on some activities Joint TAG chairs meeting 2016, Omaha Dr. Jens-Ove Heckel Zoo Landau in der Pfalz, Germany EAZA Antelope & Giraffe TAG chair [email protected] EAZA Antelope & Giraffe TAG • Antelope & Giraffe TAG continues as one of the largest and most complex TAGs representing 50 species (and approximately 90 taxa) in EAZA zoos • our remit: try to retain as many species as possible in healthy populations in EAZA collections • currently the TAG holds 11 EEPs and 11 ESBs; 4 species are part of ISBs; the remaining species are monitored (Mon-P, Mon-TAG). Chair Jens-Ove Heckel Landau Vice chair Sander Hofman Antwerp Vice chair (till 11.2015) Tania Gilbert Marwell Vice chair (since 12.2015) Kim Skalborg Simonsen Givskud Sub-group Okapi & Giraffe SG Sander Hofman Antwerp Sub-group Woodland antelope SG Kim Skalborg Simonsen Givskud Sub-group Savannah antelope SG Catrin Hammer Goerlitz Sub-group Aridlands antelope SG Ian Goodwin Marwell (Sub-group) (Mini antelope SG) (Klaus Müller-Schilling) (Hanover) Studbook keepers/Program managers EAZA Antelope & Giraffe TAG meeting 2015, Wroclaw Coordinator Conservation Tania Gilbert Marwell Coordinator Research Eulalia Moreno Almeria Advisor Veterinary Sven Hammer Goerlitz Advisor Genetics Rob Ogden Edinburgh Advisor Population management Laurie Bingaman Lackey USA Advisor Nutrition Marcus Clauss Zuerich Advisor Husbandry n.n. Advisor Education n.n. Regular support and advice also provided by: Exec. Coordinator - Collect. Coordin. & Conserv. Merel Zimmermann EPMAG / Population Management Kristin Leus Population Biologist - Collect. Coordin. & Conserv. Kristine Schad Good links to: IUCN SSC Antelope SG, IUCN Giraffe & Okapi SG/Giraffe Conservation Fdn., other EAZA Ungulate TAGs, BIAZA Hoof stock focus group, AZA Antelope & Giraffe TAG, Sahara Conservation Fund/SSIG etc. -

Common Eland
Tragelaphus oryx – Common Eland recognised, though their validity has been in dispute (Thouless 2013): Tragelaphus o. livingstonii (Sclater 1864; Livingstone's Eland): also called kaufmanni, niediecki, selousi and triangularis. It is found in the Central Zambezian Miombo woodlands i.e. south- central Africa (Angola, Zambia, Democratic Republic of the Congo, Zimbabwe, Mozambique and Malawi). Livingstone's Eland has a brown pelt with up to twelve stripes. Tragelaphus o. oryx (Pallas 1766; Cape Eland): also called alces, barbatus, canna and oreas. This subspecies is found south of the Zambezi river (South Africa, Botswana and Namibia). The fur is tawny, and adults lose their stripes. Regional Red List status (2016) Least Concern Tragelaphus o. pattersonianus (Lydekker 1906; East National Red List status (2004) Least Concern African Eland or Patterson's Eland): also called Reasons for change No change billingae. It is found in east Africa extending into the Somali arid areas, hence its common name. Its coat Global Red List status (2008) Least concern can have up to 12 stripes. TOPS listing (NEMBA) None Tragelaphus o. oryx occurs throughout the larger part of South Africa, but the far northern Limpopo Province CITES listing None bordering Zimbabwe is regarded as a transitional zone Endemic No between T. o. oryx and T. o. livingstonii or an area where they overlap. This argues the case that they should rather During drought conditions Eland roam extensively be described as ecotypes (in ecotypes, it is common for in order to meet forage and water requirements; in continuous, gradual geographic variation to impose the southern Kalahari during abnormally dry analogous phenotypic and/or genetic variation; this conditions, Eland were found to cover more than situation is called cline.). -

Copy of Roan Antelope Project
Roan Antelope Project Roan Antelope (Hippotragus equines) are Africa’s second largest antelope after the eland. Historically they were widely distributed in small herds over a large part of Swaziland. They became locally extinct when Swaziland’s very last freeranging roan antelope was poached at Maphiveni in 1961. Roan have become extremely rare across their entire natural range. The reason for their demise is probably a combination of factors which include poaching, diminishing range, stressinduced disease, predation, and competition by high density species. A small group of 4 roan (3 females, 1 bull) reintroduced to Mkhaya from Namibia perished because, we now believe, the animals were naïve to tickborne diseases in a densely tick hostile environment where mortalities overwhelmed recruitment. It is likely that the tickborne Theileria hippotragi disease played a large role. Theileria is a protozoal disease which is fatal to roan and sable antelope. The expertise of Dr Johan Steyl under the supervision of Prof. Leon Prozesky of the Onderstepoort Veterinary Faculty of the University of Pretoria has helped enormously with this project. The vector is the redlegged tick Rhipicephalus evertsi, which is abundant in Swaziland, but little was known about its impact on wildlife at the time of the Namibian introduction. Our Roan Antelope project got underway in 2002 after Dr Hamish Currie, founder and chief executive of Back to Africa, very generously committed to assist in restoring a viable population of roan antelope to Swaziland. Dr Hamish Currie wisely harnessed the expertise of Dr Johan Steyl whose guidance has without doubt been pivotal to the success of the project so far. -

Transboundary Species Project
TRANSBOUNDARY SPECIES PROJECT ROAN, SABLE AND TSESSEBE Rowan B. Martin Species Report for Roan, Sable and Tsessebe in support of The Transboundary Mammal Project of the Ministry of Environment and Tourism, Namibia facilitated by The Namibia Nature Foundation and World Wildlife Fund Living in a Finite Environment (LIFE) Programme Cover picture adapted from the illustrations by Clare Abbott in The Mammals of the Southern African Subregion by Reay H.N. Smithers Published by the University of Pretoria Republic of South Africa 1983 Transboundary Species Project – Background Study Roan, Sable and Tsessebe CONTENTS 1. BIOLOGICAL INFORMATION ...................................... 1 a. Taxonomy ..................................................... 1 b. Physical description .............................................. 3 c. Habitat ....................................................... 6 d. Reproduction and Population Dynamics ............................. 12 e. Distribution ................................................... 14 f. Numbers ..................................................... 24 g. Behaviour .................................................... 38 h. Limiting Factors ............................................... 40 2. SIGNIFICANCE OF THE THREE SPECIES ........................... 43 a. Conservation Significance ........................................ 43 b. Economic Significance ........................................... 44 3. STAKEHOLDING ................................................. 48 a. Stakeholders ................................................. -

Mixed-Species Exhibits with Pigs (Suidae)
Mixed-species exhibits with Pigs (Suidae) Written by KRISZTIÁN SVÁBIK Team Leader, Toni’s Zoo, Rothenburg, Luzern, Switzerland Email: [email protected] 9th May 2021 Cover photo © Krisztián Svábik Mixed-species exhibits with Pigs (Suidae) 1 CONTENTS INTRODUCTION ........................................................................................................... 3 Use of space and enclosure furnishings ................................................................... 3 Feeding ..................................................................................................................... 3 Breeding ................................................................................................................... 4 Choice of species and individuals ............................................................................ 4 List of mixed-species exhibits involving Suids ........................................................ 5 LIST OF SPECIES COMBINATIONS – SUIDAE .......................................................... 6 Sulawesi Babirusa, Babyrousa celebensis ...............................................................7 Common Warthog, Phacochoerus africanus ......................................................... 8 Giant Forest Hog, Hylochoerus meinertzhageni ..................................................10 Bushpig, Potamochoerus larvatus ........................................................................ 11 Red River Hog, Potamochoerus porcus ............................................................... -
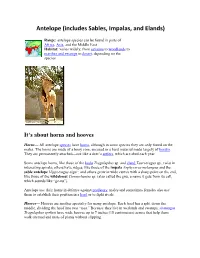
Antelope (Includes Sables, Impalas, and Elands)
Antelope (includes Sables, Impalas, and Elands) Range: antelope species can be found in parts of Africa, Asia, and the Middle East Habitat: varies widely, from savanna to woodlands to marshes and swamps to desert, depending on the species It’s about horns and hooves Horns— All antelope species have horns, although in some species they are only found on the males. The horns are made of a bony core, encased in a hard material made largely of keratin. They are permanently attached—not like a deer’s antlers, which are shed each year. Some antelope horns, like those of the kudu Tragelaphus sp. and eland Taurotragus sp., twist in interesting spirals; others have ridges, like those of the impala Aephyceros melampus and the sable antelope Hippotragus niger; and others grow in wide curves with a sharp point on the end, like those of the wildebeest Connochaetes sp. (also called the gnu, a name it gets from its call, which sounds like “ge-nu”). Antelope use their horns in defense against predators; males and sometimes females also use them to establish their position in a herd or to fight rivals Hooves— Hooves are another specialty for many antelope. Each hoof has a split down the middle, dividing the hoof into two “toes.” Because they live in wetlands and swamps, sitatungas Tragelaphus spekeii have wide hooves up to 7 inches (18 centimeters) across that help them walk on mud and mats of plants without slipping. Nile lechwes Kobus magaceros, which also live in swampy areas, have long, pointed hooves to give them sure footing in the water. -

Africa Maximum Safaris Specials 2020
AFRICA MAXIMUM SAFARIS SPECIALS 2020 South Africa North West Province Experience South Africa Package For the first time hunter to Africa we have the best package at the best price. Come experience the South African rich wildlife on a hunting safari with Africa Maximum Safaris. This is an 8-day hunt. 6 hunting days and arrival and departure day. Daily rate for 1 hunter: Cost $1600.00 Species included: Blue/Black Wildebeest or Zebra $1200 Impala $450 Springbuck $500 $450 Blesbuck Total cost $ 4200 Classic South Africa Plaines game safari 10-Day safari: 10 hunting days and arrival and departure day. Daily rates for 1 hunter and 1 observer included. Cost $ 2000.00. Species included: Kudu under 55 $2900 Gemsbuck $1,500 Blue or Black Wildebeest $1200 Impala or Blesbuck $450 Springbuck $500 Total cost $ 8550 Premier South African plainesgame safari 12 Day safaris: 10 hunting days and arrival and departure day. Daily rate for 1 hunter and 1 observer included. Cost $ 2200.00. Species Included: Greater Kudu under 55 $2,900 Cape Eland $2,900 Nyala $2,600 Kalahari Gemsbuck $1,500 Burchell Zebra $1200 Southern Impala $450 South Africa Springbuck $500 Total cost $ 14250.00 Super saver safari package 2019 Come experience a safari in South Africa at a very affordable rate. 7 Day safaris for 1 hunter and 1 observer; includes 5 hunting days and arrival and departure days. Species included in the package: Impala, Blesbuck and Blue Wildebeest. Terms and conditions: availability limited to certain months, no exchange of species or days from this package, Limited availability of packages. -

The U.K. Hunter Who Has Shot More Wildlife Than the Killer of Cecil the Lion
CAMPAIGN TO BAN TROPHY HUNTING Special Report The U.K. hunter who has shot more wildlife than the killer of Cecil the Lion SUMMARY The Campaign to Ban Trophy Hunting is revealing the identity of a British man who has killed wild animals in 5 continents, and is considered to be among the world’s ‘elite’ in the global trophy hunting industry. Malcolm W King has won a staggering 36 top awards with Safari Club International (SCI), and has at least 125 entries in SCI’s Records Book. The combined number of animals required for the awards won by King is 528. Among his awards are prizes for shooting African ‘Big Game’, wild cats, and bears. King has also shot wild sheep, goats, deer and oxen around the world. His exploits have taken him to Asia, Africa and the South Pacific, as well as across Europe. The Campaign to Ban Trophy Hunting estimates that around 1.7 million animals have been killed by trophy hunters over the past decade, of which over 200,000 were endangered species. Lions are among those species that could be pushed to extinction by trophy hunting. An estimated 10,000 lions have been killed by ‘recreational’ hunters in the last decade. Latest estimates for the African lion population put numbers at around 20,000, with some saying they could be as low as 13,000. Industry groups like Safari Club International promote prizes which actively encourage hunters to kill huge numbers of endangered animals. The Campaign to Ban Trophy Hunting believes that trophy hunting is an aberration in a civilised society. -

DRAFT SPECIES MANAGEMENT PLAN Roan Antelope Sable Antelope Tsessebe
DRAFT Ministry of Environment and Tourism Republic of Namibia SPECIES MANAGEMENT PLAN Roan antelope Hippotragus equinus Sable antelope Hippotragus niger niger Tsessebe Damaliscus lunatus lunatus May 2003 DRAFT SPECIES MANAGEMENT PLAN Roan antelope Sable antelope Tsessebe Hippotragus equinus Hippotragus niger niger Damaliscus lunatus lunatus CONTENTS GLOSSARY AND DEFINITION OF TERMS ................................... (ii) ACKNOWLEDGMENTS ................................................... (ii) EXECUTIVE SUMMARY ..................................................(iii) INTRODUCTION & BACKGROUND ............................................ 1 Conservation Status and Significance ........................................ 1 Populations ........................................................... 3 Limiting Factors and Threats .............................................. 3 Background and Rationale for the Management Plan ............................ 5 Plan Structure ......................................................... 6 MANAGEMENT PLAN ........................................................ 7 VISION AND OBJECTIVES ................................................. 7 1. Ecological Objective .................................................... 7 1.1. Strategy .......................................................... 7 1.2. Management ..................................................... 10 2. Economic Objective .................................................... 14 RISKS AND ASSUMPTIONS ............................................... 17 -

Abiotic and Anthropogenic Factors Affecting the Distribution of Four Sympatric Large Herbivores in the Mole National Park, Ghana
VOL.Ghana 59 J. Sci. 59 (2018), 23 - 30 GHANA JOURNAL OF SCIENCE https://dx.doi.org/10.4314/gjs.v59i1.223 ABIOTIC AND ANTHROPOGENIC FACTORS AFFECTING THE DISTRIBUTION OF FOUR SYMPATRIC LARGE HERBIVORES IN THE MOLE NATIONAL PARK, GHANA K. B. DAKWA Department of Conservation Biology and Entomology, School of Biological Sciences, University of Cape Coast, Ghana Email: [email protected] /[email protected] Abstract The impact of abiotic and anthropogenic factors on the distributions of buffalo (Syncerus caffer), harte- beest (Alcelaphus buselaphus), roan antelope (Hippotragus equinus) and waterbuck (Kobus defassa) at Mole National Park was assessed by transect survey. Generalized linear mixed effects logistic re- gression was used to model mammal presence/absence as a function of ecological factors. Hartebeest inhabited highlands and avoided floodplains but buffalo and roan avoided floodplains by selecting both lowlands and highlands while waterbuck inhabited lowlands but not necessarily the floodplains. Fire, water availability and anthropogenic activities were limiting factors, which constrained habitat use to make some areas unexplored for foraging. Buffalo, roan and hartebeest did not inhabit areas close to the park’s boundaries. Herbivores need optimal environment almost free of constraints to construct their distribution patterns. Therefore, management should address the problems identified in this study to ensure the herbivores’ redistribution to maximise their use of resources for their effective conservation. Introduction data on distribution of wildlife in a reserve, Mole National Park (MNP) was established conservation planning will remain inefficient in 1958 for the protection and conservation of and ineffective (Bauer et al., 2010). If we are representative savanna fauna in the northern to manage landscapes to conserve wildlife, part of Ghana. -

Antelopes, Gazelles, Cattle, Goats, Sheep, and Relatives
© Copyright, Princeton University Press. No part of this book may be distributed, posted, or reproduced in any form by digital or mechanical means without prior written permission of the publisher. INTRODUCTION RECOGNITION The family Bovidae, which includes Antelopes, Cattle, Duikers, Gazelles, Goats, and Sheep, is the largest family within Artiodactyla and the most diverse family of ungulates, with more than 270 recent species. Their common characteristic is their unbranched, non-deciduous horns. Bovids are primarily Old World in their distribution, although a few species are found in North America. The name antelope is often used to describe many members of this family, but it is not a definable, taxonomically based term. Shape, size, and color: Bovids encompass an extremely wide size range, from the minuscule Royal Antelope and the Dik-diks, weighing as little as 2 kg and standing 25 to 35 cm at the shoulder, to the Asian Wild Water Buffalo, which weighs as much as 1,200 kg, and the Gaur, which measures up to 220 cm at the shoulder. Body shape varies from relatively small, slender-limbed, and thin-necked species such as the Gazelles to the massive, stocky wild cattle (fig. 1). The forequarters may be larger than the hind, or the reverse, as in smaller species inhabiting dense tropical forests (e.g., Duikers). There is also a great variety in body coloration, although most species are some shade of brown. It can consist of a solid shade, or a patterned pelage. Antelopes that rely on concealment to avoid predators are cryptically colored. The stripes and blotches seen on the hides of Bushbuck, Bongo, and Kudu also function as camouflage by helping to disrupt the animals’ outline.