Presence and Absence of COX8 in Reptile Transcriptomes
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Biodiversity of the Southern Rupununi Savannah World Wildlife Fund and Global Wildlife Conservation
THIS REPORT HAS BEEN PRODUCED IN GUIANAS COLLABORATION VERZICHT APERWITH: Ç 2016 Biodiversity of the Southern Rupununi Savannah World Wildlife Fund and Global Wildlife Conservation 2016 WWF-Guianas Global Wildlife Conservation Guyana Office PO Box 129 285 Irving Street, Queenstown Austin, TX 78767 USA Georgetown, Guyana [email protected] www.wwfguianas.org [email protected] Text: Juliana Persaud, WWF-Guianas, Guyana Office Concept: Francesca Masoero, WWF-Guianas, Guyana Office Design: Sita Sugrim for Kriti Review: Brian O’Shea, Deirdre Jaferally and Indranee Roopsind Map: Oronde Drakes Front cover photos (left to right): Rupununi Savannah © Zach Montes, Giant Ant Eater © Gerard Perreira, Red Siskin © Meshach Pierre, Jaguar © Evi Paemelaere. Inside cover photo: Gallery Forest © Andrew Snyder. OF BIODIVERSITYTHE SOUTHERN RUPUNUNI SAVANNAH. Guyana-South America. World Wildlife Fund and Global Wildlife Conservation 2016 This booklet has been produced and published thanks to: 1 WWF Biodiversity Assessment Team Expedition Southern Rupununi - Guyana. The Southern Rupununi Biodiversity Survey Team / © WWF - GWC. Biodiversity Assessment Team (BAT) Survey. This programme was created by WWF-Guianas in 2013 to contribute to sound land- use planning by filling biodiversity data gaps in critical areas in the Guianas. As far as possible, it also attempts to understand the local context of biodiversity use and the potential threats in order to recommend holistic conservation strategies. The programme brings together local knowledge experts and international scientists to assess priority areas. With each BAT Survey, species new to science or new country records are being discovered. This booklet acknowledges the findings of a BAT Survey carried out during October-November 2013 in the southern Rupununi savannah, at two locations: Kusad Mountain and Parabara. -

Ancestral Reconstruction of Diet and Fang Condition in the Lamprophiidae: Implications for the Evolution of Venom Systems in Snakes
Journal of Herpetology, Vol. 55, No. 1, 1–10, 2021 Copyright 2021 Society for the Study of Amphibians and Reptiles Ancestral Reconstruction of Diet and Fang Condition in the Lamprophiidae: Implications for the Evolution of Venom Systems in Snakes 1,2 1 1 HIRAL NAIK, MIMMIE M. KGADITSE, AND GRAHAM J. ALEXANDER 1School of Animal, Plant and Environmental Sciences, University of the Witwatersrand, Johannesburg. PO Wits, 2050, Gauteng, South Africa ABSTRACT.—The Colubroidea includes all venomous and some nonvenomous snakes, many of which have extraordinary dental morphology and functional capabilities. It has been proposed that the ancestral condition of the Colubroidea is venomous with tubular fangs. The venom system includes the production of venomous secretions by labial glands in the mouth and usually includes fangs for effective delivery of venom. Despite significant research on the evolution of the venom system in snakes, limited research exists on the driving forces for different fang and dental morphology at a broader phylogenetic scale. We assessed the patterns of fang and dental condition in the Lamprophiidae, a speciose family of advanced snakes within the Colubroidea, and we related fang and dental condition to diet. The Lamprophiidae is the only snake family that includes front-fanged, rear-fanged, and fangless species. We produced an ancestral reconstruction for the family and investigated the pattern of diet and fangs within the clade. We concluded that the ancestral lamprophiid was most likely rear-fanged and that the shift in dental morphology was associated with changes in diet. This pattern indicates that fang loss, and probably venom loss, has occurred multiple times within the Lamprophiidae. -

Care Guide Red Eyed Crocodile Skink
Tribolonotus gracilis Care Red Eyed Guide Crocodile Skink Shy, prehistoric pets. A relatively small skink with a prehistoric appearance, the red-eyed crocodile skink is easy to care for for experienced reptile parents. While they are not incredibly active, their elusive nature and marvelous appearance are sure to inspire any reptile enthusiast fortunate enough to observe them. They do best in warm, humid, bioactive enclosures. If you are looking to add a little dinosaur to your latest terrarium build, give the red-eyed crocodile skink a try! Lifespan With proper care, these skinks have an average lifespan of 6-10+ years. Size Hatchlings measure 6-8 cm. Adults measure 6-10 inches, snout to tail. Juveniles have the same body type and skin as adults but differ in their eye and head coloration. The head of a juvenile will be cream colored and will lack the distinctive orange/red around their eyes. At around 6 months of age, the cream color will disappear, and they will develop the orange/red around their eyes. These skinks do not reach sexual maturity until 3-4 years of age, and as they mature, they develop an orange colored throat. The male is often larger than the female, but that is not always the best indicator of sex. Males have gray-blue raised pores on the underside of the third and fourth toes of their hind feet. Males also have a rectangular section of four to six enlarged belly scales at the umbilicus. Natural History There are eight known species in the genus Tribolonotus, which are all extant to Irian Jaya and Papua New Guinea in the Solomon Islands. -

DESDE EL OJO DE LA CÁMARA TRAMPA Mamíferos, Aves Y Reptiles Del Río La Novia (Purús, Perú)
DESDE EL OJO DE LA CÁMARA TRAMPA Mamíferos, aves y reptiles del río La Novia (Purús, Perú) Hiromi Yagui Briones Heidi Rubio Torgler Jose Luis Mena Álvarez Cómo citar este libro: Hiromi Yagui Briones, Heidi Rubio Torgler, José Luis Mena Álvarez (2015). Desde el ojo de la cámara trampa. Consorcio Purús-Manu: WWF, CARE Perú, ProNaturaleza, ProPurús, Sociedad Zoológica de Fráncfort, ORAU. Lima, noviembre de 2015. Primera edición: 2015 © WWF Perú Razón social: World Wildlife Fund Inc. Domicilio: Av. Trinidad Morán 853, Lince Teléfono: (511) 440-5550 Impreso en Ediciones Nova Print S.A.C. Av. Ignacio Merino 1546 Lince 1000 ejemplares Autores: Hiromi Yagui Briones, Heidi Rubio Torgler, Jose Luis Mena Álvarez Fotos: WWF Perú Dibujos a tinta: Felipe Morey Gamarra Gráficos: Hiromi Yagui Briones Edición de mapas: Alejandro Tello Martínez, Hiromi Yagui Briones Edición de estilo: Jhonathan Jara Giudiche Diagramación: Jorge Kajatt Producido en Perú, 2015 Esta publicación ha sido posible gracias al apoyo del Pueblo de los Estados Unidos de América a través de la Agencia de los Estados Unidos para el Desarrollo Internacional (USAID). Las opiniones aquí expresadas son las del autor(es) y no reflejan necesariamente la opinión de USAID ni del Gobierno de los Estados Unidos. Agradecimientos Un agradecimiento especial a los miembros de MABOSINFRON; al señor Carlos Loja Manuyama e Isaías Pérez Ramírez, quienes dedicaron todo su tiempo para la ejecución de las actividades en campo, quienes fueron grandes guías en campo que también llegaron a manejar de manera óptima las cámaras instaladas y a los señores Amancio Flores Lomas (en ese entonces presidente de la organización) y a Miguel Ruiz Pérez por la ayuda en todo momento para que la investigación se pueda desarrollar sin ningún contratiempo. -

New Verified Nonindigenous Amphibians and Reptiles in Florida Through 2015, with a Summary of Over 152 Years of Introductions
WWW.IRCF.ORG/REPTILESANDAMPHIBIANSJOURNALTABLE OF CONTENTS IRCF REPTILES & IRCF AMPHIBIANS REPTILES • VOL &15, AMPHIBIANS NO 4 • DEC 2008 • 189 23(2):110–143 • AUG 2016 IRCF REPTILES & AMPHIBIANS CONSERVATION AND NATURAL HISTORY TABLE OF CONTENTS INTRODUCED SPECIES FEATURE ARTICLES . Chasing Bullsnakes (Pituophis catenifer sayi) in Wisconsin: New VerifiedOn the Road to Understanding the Nonindigenous Ecology and Conservation of the Midwest’s Giant Serpent ...................... Amphibians Joshua M. Kapfer 190 . The Shared History of Treeboas (Corallus grenadensis) and Humans on Grenada: A Hypothetical Excursion ............................................................................................................................Robert W. Henderson 198 and ReptilesRESEARCH ARTICLES in Florida through 2015, with a . The Texas Horned Lizard in Central and Western Texas ....................... Emily Henry, Jason Brewer, Krista Mougey, and Gad Perry 204 Summary. The Knight Anole of(Anolis equestris over) in Florida 152 Years of Introductions .............................................Brian J. Camposano, Kenneth L. Krysko, Kevin M. Enge, Ellen M. Donlan, and Michael Granatosky 212 1 1 2 3 3 4 Kenneth L. KryskoCONSERVATION, Louis A. Somma ALERT, Dustin C. Smith , Christopher R. Gillette , Daniel Cueva , Joseph A. Wasilewski , 5 6 7 8 9 10 Kevin M. Enge. , Steve A. Johnson , Todd S. Campbell , Jake R. Edwards , Michael R. Rochford , Rhyan Tompkins , World’s Mammals11 in Crisis .............................................................................................................................................................12 -

Systematics of the Carlia “Fusca” Lizards (Squamata: Scincidae) of New Guinea and Nearby Islands
Systematics of the Carlia “fusca” Lizards (Squamata: Scincidae) of New Guinea and Nearby Islands George R. Zug Bishop Museum Bulletin in Zoology 5 Bishop Museum Press Honolulu, 2004 Cover: Published by Bishop Museum Press 1525 Bernice Street Honolulu, Hawai‘i 96817-2704, USA Copyright ©2004 Bishop Museum All Rights Reserved Printed in the United States of America ISSN 0893-312X Zug — Carlia “fusca” Lizards from New Guinea and Nearby Islands v TABLE OF CONTENTS Acknowledgments ......................................................................................................................... vii Abstract ........................................................................................................................................ viii Introduction ................................................................................................................................... 1 Carlia: An Analysis for Species Relationships ........................................................................... 1 Characters and Taxa .................................................................................................................. 2 Phylogenetic Analysis................................................................................................................ 8 New Guinea Carlia “fusca” ....................................................................................................... 9 Materials and Methods................................................................................................................. -
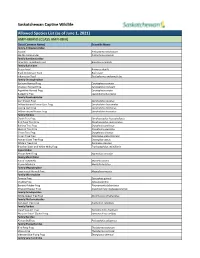
Captive Wildlife Allowed List
Saskatchewan Captive Wildlife Allowed Species List (as of June 1, 2021) AMPHIBIANS (CLASS AMPHIBIA) Class (Common Name) Scientific Name Family Ambystomatidae Axolotl Ambystoma mexicanum Marble Salamander Ambystoma opacum Family Bombinatoridae Oriental Fire-Bellied Toad Bombina orientalis Family Bufonidae Green Toad Anaxyrus debilis Black Indonesian Toad Bufo asper Indonesian Toad Duttaphrynus melanostictus Family Ceratophryidae Surinam Horned Frog Ceratophrys cornuta Chacoan Horned Frog Ceratophrys cranwelli Argentine Horned Frog Ceratophrys ornata Budgett’s Frog Lepidobatrachus laevis Family Dendrobatidae Dart Poison Frog Dendrobates auratus Yellow-banded Poison Dart Frog Dendrobates leucomelas Dyeing Dart Frog Dendrobates tinctorius Yellow-striped Poison Frog Dendrobates truncatus Family Hylidae Clown Tree Frog Dendropsophus leucophyllatus Bird Poop Tree Frog Dendropsophus marmoratus Barking Tree Frog Dryophytes gratiosus Squirrel Tree Frog Dryophytes squirellus Green Tree Frog Dryophytes cinereus Cuban Tree Frog Osteopilus septentrionalis Haitian Giant Tree Frog Osteopilus vastus White’s Tree Frog Ranoidea caerulea Brazilian Black and White Milky Frog Trachycephalus resinifictrix Hyperoliidae African Reed Frog Hyperolius concolor Family Mantellidae Baron’s Mantella Mantella baroni Brown Mantella Mantella betsileo Family Megophryidae Long-nosed Horned Frog Megophrys nasuta Family Microhylidae Tomato Frog Dyscophus guineti Chubby Frog Kaloula pulchra Banded Rubber Frog Phrynomantis bifasciatus Emerald Hopper Frog Scaphiophryne madagascariensis -

Species Richness in Time and Space: a Phylogenetic and Geographic Perspective
Species Richness in Time and Space: a Phylogenetic and Geographic Perspective by Pascal Olivier Title A dissertation submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy (Ecology and Evolutionary Biology) in The University of Michigan 2018 Doctoral Committee: Assistant Professor and Assistant Curator Daniel Rabosky, Chair Associate Professor Johannes Foufopoulos Professor L. Lacey Knowles Assistant Professor Stephen A. Smith Pascal O Title [email protected] ORCID iD: 0000-0002-6316-0736 c Pascal O Title 2018 DEDICATION To Judge Julius Title, for always encouraging me to be inquisitive. ii ACKNOWLEDGEMENTS The research presented in this dissertation has been supported by a number of research grants from the University of Michigan and from academic societies. I thank the Society of Systematic Biologists, the Society for the Study of Evolution, and the Herpetologists League for supporting my work. I am also extremely grateful to the Rackham Graduate School, the University of Michigan Museum of Zoology C.F. Walker and Hinsdale scholarships, as well as to the Department of Ecology and Evolutionary Biology Block grants, for generously providing support throughout my PhD. Much of this research was also made possible by a Rackham Predoctoral Fellowship, and by a fellowship from the Michigan Institute for Computational Discovery and Engineering. First and foremost, I would like to thank my advisor, Dr. Dan Rabosky, for taking me on as one of his first graduate students. I have learned a tremendous amount under his guidance, and conducting research with him has been both exhilarating and inspiring. I am also grateful for his friendship and company, both in Ann Arbor and especially in the field, which have produced experiences that I will never forget. -

The Evolution of Diet in the Lamprophiidae Hiral Naik 452805
The evolution of diet in the Lamprophiidae Hiral Naik 452805 A Dissertation submitted to the School of Animal, Plant and Environmental Sciences, University of the Witwatersrand, Johannesburg, South Africa in fulfilment of the requirements of the degree of Master of Science. Johannesburg, South Africa July 2017 Declaration I declare that this dissertation is my own, unaided work unless specifically acknowledged in the text. It has not been submitted previously for any degree or examination at any other university, nor has it been prepared under the aegis or with the assistance of any other body or organization or person outside of the University of the Witwatersrand, Johannesburg, South Africa. ______________ Hiral Naik 13/07/2017 1 Abstract Studying feeding biology in a phylogenetic context helps elucidate the factors that significantly influenced the evolutionary history of organisms. The snake lineage is one of the most morphologically and ecologically diverse clades of vertebrates due to a variety of traits (e.g. venom, body shape, gape size and habitat use) that have enabled their exceptional radiation. Recently, the Deep History Hypothesis (DHH) has been used to explain how divergence, deep in the evolutionary history of snakes, has resulted in present day niche preferences. The Competition-Predation Hypothesis (CPH) contrastingly attributes current ecological traits to recent species interactions. Diet has been a key factor in shaping snake diversity and ecology, and it has often been used as a proxy to understand current snake community structure and evolutionary trends in snakes. I tested the validity of the two evolutionary hypotheses in the Lamprophiidae, a family of primarily African snakes. -
And “Exotic” Meats
See discussions, stats, and author profiles for this publication at: https://www.researchgate.net/publication/301759777 Controversial cuisine: A global account of the demand, supply and acceptance of “unconventional” and “exotic” meats Article in Meat Science · April 2016 DOI: 10.1016/j.meatsci.2016.04.017 CITATIONS READS 0 155 2 authors: Donna-Mareè Cawthorn Louwrens C Hoffman University of Salford Stellenbosch University 28 PUBLICATIONS 335 CITATIONS 276 PUBLICATIONS 2,476 CITATIONS SEE PROFILE SEE PROFILE All in-text references underlined in blue are linked to publications on ResearchGate, Available from: Donna-Mareè Cawthorn letting you access and read them immediately. Retrieved on: 17 September 2016 MESC-06976; No of Pages 18 Meat Science xxx (2016) xxx–xxx Contents lists available at ScienceDirect Meat Science journal homepage: www.elsevier.com/locate/meatsci Controversial cuisine: A global account of the demand, supply and acceptance of “unconventional” and “exotic” meats Donna-Mareè Cawthorn, Louwrens C. Hoffman ⁎,1 Department of Animal Sciences, University of Stellenbosch, Private Bag X1, Matieland 7600, South Africa article info abstract Article history: In most societies, meat is more highly prized, yet more frequently tabooed, than any other food. The reasons for Received 18 January 2016 these taboos are complex and their origins have been the focus of considerable research. In this paper, we illus- Received in revised form 6 April 2016 trate this complexity by deliberating on several “unconventional” or “exotic” animals that are eaten around the Accepted 11 April 2016 world, but whose consumption evokes strong emotions, controversy and even national discourse: dogs, equids, Available online xxxx kangaroos, marine mammals, primates, rodents and reptiles. -

Amphibian and Reptile Records from Around the Betsiboka Delta Area in North-Western Madagascar
Herpetology Notes, volume 8: 535-543 (2015) (published online on 24 November 2015) Amphibian and reptile records from around the Betsiboka delta area in North-Western Madagascar Andolalao Rakotoarison1, 2,*, Jesse Erens1, 3, Fanomezana M. Ratsoavina2 and Miguel Vences1 Abstract. This study summarises amphibian and reptile records from ad hoc surveys in a series of localities in the North-West of Madagascar, largely centred on the delta of the Betsiboka River. Eleven amphibian and approximately 32 reptile species were found, with taxonomic uncertainties remaining for some of them. Among the most relevant findings, we report on range extensions northwards of Aglyptodactylus laticeps (verified by DNA sequencing), and of an enigmatic skink of the Trachylepis aureopunctata group, possibly close to T. dumasi, T. tandrefana, or T. volamenaloha. We furthermore provide anecdotal information on habitat and natural history of several rare and regionally endemic burrowing skinks, i.e., Voeltzkowia mira, V. yamagishii, and Pygomeles petteri. Key words: Range extension, Aglyptodactylus laticeps, Trachylepis sp. aff. dumasi, natural history, Voeltzkowia mira, Pygomeles petteri. Introduction 2007) and species delimitation has been improved by comprehensive molecular data sets (Vieites et al., 2009; In hyperdiverse tropical faunas of amphibians and Nagy et al., 2012; Perl et al., 2014), the knowledge reptiles, the biology and population dynamics of the of some regions and taxa remains scarce. The dry to vast majority of species remains poorly known, and subarid regions in the South-West, West and North- information on their spatial occurrence becomes West (regions according to Boumans et al. 2007) paramount for their conservation. In Madagascar, Red contain numerous such poorly accessible and poorly List assessments (Andreone et al., 2005; Jenkins, 2015) explored sites, but at the same time are characterized by and reserve planning (Kremen et al., 2008) are largely high rates of habitat destruction (Waeber et al., 2015). -

NOTES Canopy Chameleon (Furcifer Willsii) Consumption by Common Big
NOTES Canopy chameleon (Furcifer willsii) consumption by common big-eyed snake (Mimophis mahfalensis) in Fivahona Forest, eastern Madagascar Javier Lobón-Rovira1, Francesco Belluardo1, 2012, 2016a; Cove et al., 2017; Raselimanana, Malalatiana Rasoazanany2, Gonçalo M. Rosa3,4 & 2018), and general patterns are still difficult to unveil. Angelica Crottini1,5 The Malagasy common big-eyed snake, Mimophis 1 CIBIO Centro de Investigação em Biodiversidade e mahfalensis, is a widespread species and perhaps Recursos Genéticos, Universidade do Porto, Rua Padre the most common diurnal snake encountered in the Armando Quintas, 7, 4485-661 Vairão, Portugal southwestern portion of the island. This lamprophiid E-mail: [email protected], snake is widely distributed along the central and [email protected] southern regions of the country (Nagy et al., 2003; 2 Mention Zoologie et Biodiversité Animale, Faculté Glaw & Vences, 2007; Ruane et al., 2018). Frogs, des Sciences, BP 906, Université d’Antananarivo, skinks, plated lizards, and chameleons are common Antananarivo101, Madagascar items in its diet (Jenkins et al., 2009; Rosa et al., E-mail: [email protected] 2016b; Raselimanana, 2018). Here we report on a 3 Institute of Zoology, Zoological Society of London, new prey item to Mimophis mahfalensis, based on the Regent’s Park, NW1 4RY London, United Kingdom inspection of stomach contents. E-mail: [email protected] 4 Centre for Ecology, Evolution and Environmental During a herpetological survey in the areas Changes (cE3c), Faculdade de Ciências da surrounding the Andringitra Massif (Haute Matsiatra Universidade de Lisboa, Edifício C2, 5º Piso, Sala Region), we investigated Velotsoa Forest, a remote 2.5.46 Campo Grande, 1749-016 Lisboa, Portugal fragment of humid forest close to the village of 5 Departamento de Biologia, Faculdade de Ciências da Fivahona.