Zebrafish Models of Neurodevelopmental
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Role of Citicoline in the Management of Traumatic Brain Injury
pharmaceuticals Review Role of Citicoline in the Management of Traumatic Brain Injury Julio J. Secades Medical Department, Ferrer, 08029 Barcelona, Spain; [email protected] Abstract: Head injury is among the most devastating types of injury, specifically called Traumatic Brain Injury (TBI). There is a need to diminish the morbidity related with TBI and to improve the outcome of patients suffering TBI. Among the improvements in the treatment of TBI, neuroprotection is one of the upcoming improvements. Citicoline has been used in the management of brain ischemia related disorders, such as TBI. Citicoline has biochemical, pharmacological, and pharmacokinetic characteristics that make it a potentially useful neuroprotective drug for the management of TBI. A short review of these characteristics is included in this paper. Moreover, a narrative review of almost all the published or communicated studies performed with this drug in the management of patients with head injury is included. Based on the results obtained in these clinical studies, it is possible to conclude that citicoline is able to accelerate the recovery of consciousness and to improve the outcome of this kind of patient, with an excellent safety profile. Thus, citicoline could have a potential role in the management of TBI. Keywords: CDP-choline; citicoline; pharmacological neuroprotection; brain ischemia; traumatic brain injury; head injury Citation: Secades, J.J. Role of 1. Introduction Citicoline in the Management of Traumatic brain injury (TBI) is among the most devastating types of injury and can Traumatic Brain Injury. result in a different profile of neurological and cognitive deficits, and even death in the most Pharmaceuticals 2021, 14, 410. -

Drug-Excipient Interaction and Its Importance in Dosage Form
Journal of Applied Pharmaceutical Science 01 (06); 2011: 66-71 ISSN: 2231-3354 Drug-excipient interacti on and its importance in Received: 29-07-2011 Revised on: 02-08-2011 Accepted: 04-08-2011 dosage form development Nishath Fathima, Tirunagari Mamatha, Husna Kanwal Qureshi, Nandagopal Anitha and Jangala Venkateswara Rao ABSTRACT Excipients are included in dosage forms to aid manufacture, administration or absorption. Although considered pharmacologically inert, excipients can initiate, propagate or participate in Nishath Fathima, Tirunagari chemical or physical interactions with drug compounds, which may compromise the effectiveness Mamatha, Husna Kanwal Qureshi, of a medication. Exicipients are not exquisitely pure. Even for the most commonly used excipients, Nandagopal Anitha and Jangala it is necessary to understand the context of their manufacture in order to identify potential active pharmaceutical ingredients interactions with trace components. Chemical interactions can lead to Venkateswara Rao Sultan-Ul-Uloom College of Pharmacy, degradation of the active ingredient, thereby reducing the amount available for therapeutic effect. Physical interactions can affect rate of dissolution, uniformity of dose or ease of administration. Banjara Hills, Hyderabad - 500034, A.P., India. Key words: Excipient, Drug, Interaction, Physical, Chemical. INTRODUCTION Pharmaceutical dosage form is a combination of active pharmaceutical ingredients (API) and excipients. Excipients are included in dosage forms to aid manufacture, administration or absorption (Crowley and Martini).The ideal excipients must be able to fulfill the important functions i.e. dose, stability and release of API from the formulation. Although considered pharmacologically inert, excipients can initiate, propagate or participate in chemical or physical interactions with drug compounds, which may compromise the effectiveness of a medication. -

DNA Modifications in Models of Alcohol Use Disorders
Alcohol 60 (2017) 19e30 Contents lists available at ScienceDirect Alcohol journal homepage: http://www.alcoholjournal.org/ DNA modifications in models of alcohol use disorders * Christopher T. Tulisiak a, R. Adron Harris a, b, Igor Ponomarev a, b, a Waggoner Center for Alcohol and Addiction Research, The University of Texas at Austin, 2500 Speedway, A4800, Austin, TX 78712, USA b The College of Pharmacy, The University of Texas at Austin, 2409 University Avenue, A1900, Austin, TX 78712, USA article info abstract Article history: Chronic alcohol use and abuse result in widespread changes to gene expression, some of which Received 2 September 2016 contribute to the development of alcohol-use disorders (AUD). Gene expression is controlled, in part, by a Received in revised form group of regulatory systems often referred to as epigenetic factors, which includes, among other 3 November 2016 mechanisms, chemical marks made on the histone proteins around which genomic DNA is wound to Accepted 5 November 2016 form chromatin, and on nucleotides of the DNA itself. In particular, alcohol has been shown to perturb the epigenetic machinery, leading to changes in gene expression and cellular functions characteristic of Keywords: AUD and, ultimately, to altered behavior. DNA modifications in particular are seeing increasing research Alcohol fi Epigenetics in the context of alcohol use and abuse. To date, studies of DNA modi cations in AUD have primarily fi fi fi DNA methylation looked at global methylation pro les in human brain and blood, gene-speci c methylation pro les in DNMT animal models, methylation changes associated with prenatal ethanol exposure, and the potential DNA hydroxymethylation therapeutic abilities of DNA methyltransferase inhibitors. -

A Single-Label Phenylpyrrolocytidine Provides a Molecular Beacon-Like Response Reporting HIV-1 RT Rnase H Activity Alexander S
1048–1056 Nucleic Acids Research, 2010, Vol. 38, No. 3 Published online 20 November 2009 doi:10.1093/nar/gkp1022 A single-label phenylpyrrolocytidine provides a molecular beacon-like response reporting HIV-1 RT RNase H activity Alexander S. Wahba1, Abbasali Esmaeili2, Masad J. Damha1 and Robert H. E. Hudson3,* 1Department of Chemistry, McGill University, Montreal, QC, H3A 2K6 Canada, 2Department of Chemistry, University of Birjand, Birjand, Iran and 3Department of Chemistry, University of Western Ontario, London, ON, N6A 5B7 Canada Downloaded from https://academic.oup.com/nar/article/38/3/1048/3112336 by guest on 27 September 2021 Received August 15, 2009; Revised and Accepted October 19, 2009 ABSTRACT identified as a potential target for antiretroviral therapy as it is required for virus infectivity (3); yet there are no 6-Phenylpyrrolocytidine (PhpC), a structurally con- antiRNase H agents in clinical use. Few inhibitors of HIV- servative and highly fluorescent cytidine analog, 1 RNase H were identified until the transition of testing was incorporated into oligoribonucleotides. The methods from gel-based techniques to fluorescent assays PhpC-containing RNA formed native-like duplex amenable to high-throughput screening (HTS) (4–8). The structures with complementary DNA or RNA. The most widely used assay was developed by Parniak and co- PhpC-modification was found to act as a sensitive workers (6) and utilizes a two label, molecular beacon reporter group being non-disruptive to structure and strategy (9) in which the RNA strand is labeled with a 0 the enzymatic activity of RNase H. A RNA/DNA 3 -terminal fluorophore (fluorescein, F) and a DNA 0 hybrid possessing a single PhpC insert was an strand with a quencher (dabcyl, Q) at the 5 -terminus excellent substrate for HIV-1 RT Ribonuclease H (Scheme 1). -

MR Evaluation of Hydrocephalus
591 MR Evaluation of Hydrocephalus Taher EI Gammal1 An analysis of sagittal T1-weighted MR studies was performed in 23 patients with Marshall B. Allen, Jr.2 hydrocephalus, 58 patients with atrophy, and 100 normal patients. The average rna mil Betty Sue Brooks 1 lopontine distance was 1.15 cm for the normal group, 1.2 cm for patients with atrophy, Edward K. Mark2 and 7.5 mm for patients with hydrocephalus. A reduction of the mamillopontine distance below 1.0 cm was found in 22 patients with hydrocephalus,S patients with atrophy, and 15 normal patients. Dilatation of the anterior third ventricle was noted in 21 patients in the hydrocephalus group and in none of the patients in the atrophy and normal groups. The average thickness of the corpus callosum at the level of the foramen of Monro was 6 mm in normal subjects and was reduced below 6 mm in 16 of the hydrocephalus patients. Smooth elevation of the corpus callosum was noted in 20 hydrocephalus patients, in 2 patients with atrophy, and in none of the normal patients. MR improves the accuracy of diagnosis in patients with hydrocephalus both because of its ability to show small obstructing lesions that are not depicted by CT and because the mass effect of the distended supratentorial ventricles produces anatomic changes that are delineated with precision by MR. CT provides incomplete information for diagnosing hydrocephalus produced by very small or isodense pathologic changes in the aqueduct and in the foramen of Monro [1, 2]. Even with advanced CT techniques, use of contrast media in the subarachnoid space and ventricular system was essential in the CT evaluation of some cases of hydrocephalus [3-5]. -

Polymeric Derivative of Cytidine Metabolic Antagonist Polymer-Derivat Von Cytidin Metabolit Antagonist Derive Polymere D’Un Antagoniste Metabolique De La Cytidine
(19) & (11) EP 1 881 020 B1 (12) EUROPEAN PATENT SPECIFICATION (45) Date of publication and mention (51) Int Cl.: of the grant of the patent: C08G 65/08 (2006.01) A61K 38/00 (2006.01) 11.08.2010 Bulletin 2010/32 C08G 65/333 (2006.01) C08G 69/48 (2006.01) (21) Application number: 06745754.9 (86) International application number: PCT/JP2006/308826 (22) Date of filing: 27.04.2006 (87) International publication number: WO 2006/120914 (16.11.2006 Gazette 2006/46) (54) POLYMERIC DERIVATIVE OF CYTIDINE METABOLIC ANTAGONIST POLYMER-DERIVAT VON CYTIDIN METABOLIT ANTAGONIST DERIVE POLYMERE D’UN ANTAGONISTE METABOLIQUE DE LA CYTIDINE (84) Designated Contracting States: • MASHIBA, Hiroko, AT BE BG CH CY CZ DE DK EE ES FI FR GB GR NIPPON KAYAKU KABUSHIKI KAISHA HU IE IS IT LI LT LU LV MC NL PL PT RO SE SI Tokyo 1150042 (JP) SK TR • YAMAMOTO, Keiichirou, Designated Extension States: c/o NIPPON KAYAKU K.K. AL BA HR MK YU Tokyo 1150042 (JP) • TAKASHIO, Kazutoshi, (30) Priority: 11.05.2005 JP 2005138249 NIPPON KAYAKU K.K. Tokyo 1150042 (JP) (43) Date of publication of application: 23.01.2008 Bulletin 2008/04 (74) Representative: Wablat, Wolfgang Patentanwalt (73) Proprietor: NIPPON KAYAKU KABUSHIKI KAISHA Dr. Dr. W. Wablat Tokyo 102-8172 (JP) Potsdamer Chaussee 48 14129 Berlin (DE) (72) Inventors: • MASUDA, Akira, (56) References cited: c/o NIPPON KAYAKU KABUSHIKI KAISHA EP-A- 0 097 307 EP-A- 0 685 504 Tokyo 1150042 (JP) WO-A-01/21135 WO-A-02/065988 • ONDA, Takeshi, JP-A- 6 206 832 JP-A- 02 300 133 c/o NIPPON KAYAKU KABUSHIKI KAISHA JP-A- 06 206 832 JP-A- 08 048 766 Tokyo 1150042 (JP) JP-A- 2003 524 028 JP-A- 2004 532 289 Note: Within nine months of the publication of the mention of the grant of the European patent in the European Patent Bulletin, any person may give notice to the European Patent Office of opposition to that patent, in accordance with the Implementing Regulations. -
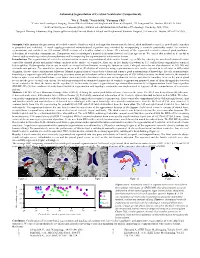
Automated Segmentation of Cerebral Ventricular Compartments
Automated Segmentation of Cerebral Ventricular Compartments 1Wu Y, 2Pohl K, 3Warfield SK, 1Cuttmann CRG. 1Center for Neurological Imaging, Harvard Medical School and Brigham and Women's Hospital, 221 Longwood Av., Boston, MA 02115 USA, 2Artificial Intelligence Laboratory,http://www.ai.mit.edu Massachusetts Institute of Technology, Cambridge MA, USA., 3Surgical Planning Laboratory http://www.spl.harvard.edu Harvard Medical School and Brigham and Women's Hospital, 75 Francis St., Boston, MA 02115 USA, Synopsis: Fully automated segmentation of cerebral ventricle chambers, which is designed to discriminate the lateral, third and fourth ventricles, as well as the aqueduct is presented and validated. A novel topology-restricted intensity-based algorithm was extended by incorporating a ventricle probability model for ventricle segmentation, and validated on 124 coronal SPGR sections of a healthy volunteer’s brain. 3D-rendering of the segmented ventricles showed good qualitative delineation of ventricular compartments. Comparison with a radiologist’s manual delineation showed excellent agreement. We expect this method to be useful in clinical studies involving ventricular morphometry and for improving the segmentation of white matter lesions. Introduction: The segmentation of ventricles is important for accurate segmentation of white matter lesions, e.g. in MS. by reducing the misclassification of lesions caused by choroid plexus and partial volume artifacts at the surface of ventricles. Also, one in five hundred newborn in U.S. suffers from congenital or acquired hydrocephalus. Hydrocephalus also occurs in adults as a result of head trauma, meningitis, tumors or cysts. Enlarged ventricles are also observed in AD, MS and schizophrenia patients. The quantitative measurement, as well as 3D display of ventricles using segmentation in vivo can be expected to be of value in differential diagnosis, disease characterization and follow-up. -

Neuroanatomy Dr
Neuroanatomy Dr. Maha ELBeltagy Assistant Professor of Anatomy Faculty of Medicine The University of Jordan 2018 Prof Yousry 10/15/17 A F B K G C H D I M E N J L Ventricular System, The Cerebrospinal Fluid, and the Blood Brain Barrier The lateral ventricle Interventricular foramen It is Y-shaped cavity in the cerebral hemisphere with the following parts: trigone 1) A central part (body): Extends from the interventricular foramen to the splenium of corpus callosum. 2) 3 horns: - Anterior horn: Lies in the frontal lobe in front of the interventricular foramen. - Posterior horn : Lies in the occipital lobe. - Inferior horn : Lies in the temporal lobe. rd It is connected to the 3 ventricle by body interventricular foramen (of Monro). Anterior Trigone (atrium): the part of the body at the horn junction of inferior and posterior horns Contains the glomus (choroid plexus tuft) calcified in adult (x-ray&CT). Interventricular foramen Relations of Body of the lateral ventricle Roof : body of the Corpus callosum Floor: body of Caudate Nucleus and body of the thalamus. Stria terminalis between thalamus and caudate. (connects between amygdala and venteral nucleus of the hypothalmus) Medial wall: Septum Pellucidum Body of the fornix (choroid fissure between fornix and thalamus (choroid plexus) Relations of lateral ventricle body Anterior horn Choroid fissure Relations of Anterior horn of the lateral ventricle Roof : genu of the Corpus callosum Floor: Head of Caudate Nucleus Medial wall: Rostrum of corpus callosum Septum Pellucidum Anterior column of the fornix Relations of Posterior horn of the lateral ventricle •Roof and lateral wall Tapetum of the corpus callosum Optic radiation lying against the tapetum in the lateral wall. -

Non-Enzymatic Synthesis of the Coenzymes, Uridine Diphosphate
N O N - E N Z Y M A T I C S Y N T H E S I S OF THE C O E N Z Y M E S , U R I D I N E D I P H O S P H A T E G L U C O S E A N D C Y T I D I N E D I P H O S P H A T E C H O L I N E , A N D O T H E R P H O S P H O R Y L A T E D M E T A B O L I C I N T E R M E D I A T E S A. M A R , J. D W O R K I N , and J. ORO* Department of Biochemical and Biophysical Sciences, University of Houston, Houston, TX 77004, U.S.A. (Received 3 November, 1986) Abstract. The synthesis of uridine diphosphate glucose (UDPG), cytidine diphosphate choline (CDP- choline), glucose-l-phosphate (G1P) and glucose-6-phosphate (G6P) has been accomplished under simulated prebiotic conditions using urea and cyanamide, two condensing agents considered to have been present on the primitive Earth. The synthesis of UDPG was carried out by reacting G1P and UTP at 70 °C for 24 hours in the presence of the condensing agents in an aqueous medium. CDP-choline was obtained under the same conditions by reacting choline phosphate and CTP. G1P and G6P were synthesized from glucose and inorganic phosphate at 70°C for 16 hours. -

Minocycline As a Candidate Treatment
Behavioural Brain Research 235 (2012) 302–317 Contents lists available at SciVerse ScienceDirect Behavioural Brain Research j ournal homepage: www.elsevier.com/locate/bbr Review Novel therapeutic targets in depression: Minocycline as a candidate treatment a,b c,d c,d f,g Joanna K. Soczynska , Rodrigo B. Mansur , Elisa Brietzke , Walter Swardfager , a,b,e b b Sidney H. Kennedy , Hanna O. Woldeyohannes , Alissa M. Powell , b a,b,e,f,∗ Marena S. Manierka , Roger S. McIntyre a Institute of Medical Science, University of Toronto, Toronto, Canada b Mood Disorders Psychopharmacology Unit, University Health Network, Toronto, Canada c Program of Recognition and Intervention in Individuals in at Risk Mental States (PRISMA), Department of Psychiatry, Universidade Federal de São Paulo, São Paulo, Brazil d Interdisciplinary Laboratory of Clinical Neurosciences (LINC), Department of Psychiatry, Universidade Federal de São Paulo, São Paulo, Brazil e Department of Psychiatry, University of Toronto, Toronto, Canada f Departments of Pharmacology and Toxicology, University of Toronto, Toronto, Canada g Neuropsychopharmacology Research Group, Sunnybrook Health Sciences Centre, Toronto, Canada h i g h l i g h t s Regional cell loss and brain atrophy in mood disorders may be a consequence of impaired neuroplasticity. Neuroplasticity is regulated by neurotrophic, inflammatory, oxidative, glutamatergic pathways. Abnormalities in these systems are implicated in the pathophysiology of mood disorders. Minocycline exerts effects on neuroplasticity and targets these interacting systems. Evidence indicates that minocycline may be a viable treatment option for mood disorders. a r t i c l e i n f o a b s t r a c t Article history: Mood disorders are marked by high rates of non-recovery, recurrence, and chronicity, which are insuf- Received 1 December 2011 ficiently addressed by current therapies. -

Customs Tariff - Schedule
CUSTOMS TARIFF - SCHEDULE 99 - i Chapter 99 SPECIAL CLASSIFICATION PROVISIONS - COMMERCIAL Notes. 1. The provisions of this Chapter are not subject to the rule of specificity in General Interpretative Rule 3 (a). 2. Goods which may be classified under the provisions of Chapter 99, if also eligible for classification under the provisions of Chapter 98, shall be classified in Chapter 98. 3. Goods may be classified under a tariff item in this Chapter and be entitled to the Most-Favoured-Nation Tariff or a preferential tariff rate of customs duty under this Chapter that applies to those goods according to the tariff treatment applicable to their country of origin only after classification under a tariff item in Chapters 1 to 97 has been determined and the conditions of any Chapter 99 provision and any applicable regulations or orders in relation thereto have been met. 4. The words and expressions used in this Chapter have the same meaning as in Chapters 1 to 97. Issued January 1, 2019 99 - 1 CUSTOMS TARIFF - SCHEDULE Tariff Unit of MFN Applicable SS Description of Goods Item Meas. Tariff Preferential Tariffs 9901.00.00 Articles and materials for use in the manufacture or repair of the Free CCCT, LDCT, GPT, UST, following to be employed in commercial fishing or the commercial MT, MUST, CIAT, CT, harvesting of marine plants: CRT, IT, NT, SLT, PT, COLT, JT, PAT, HNT, Artificial bait; KRT, CEUT, UAT, CPTPT: Free Carapace measures; Cordage, fishing lines (including marlines), rope and twine, of a circumference not exceeding 38 mm; Devices for keeping nets open; Fish hooks; Fishing nets and netting; Jiggers; Line floats; Lobster traps; Lures; Marker buoys of any material excluding wood; Net floats; Scallop drag nets; Spat collectors and collector holders; Swivels. -

Ep 0882734 A2
Patentamt Europaisches |||| ||| 1 1|| ||| ||| || || || ||| || ||| || || || (19) J European Patent Office Office europeen des brevets (11) EP 0 882 734 A2 (12) EUROPEAN PATENT APPLICATION (43) Date of publication:ation: (51) |nt. ci.6: C07H 19/167, A61 K 31/70 09.12.1998 Bulletin 1998/50 (21) Application number: 98109318.0 (22) Date of filing: 22.05.1998 (84) Designated Contracting States: • Ishikawa, Tohru AT BE CH CY DE DK ES Fl FR GB GR IE IT LI LU Kanagawa-ken (JP) MC NL PT SE • Ishitsuka, Hideo Designated Extension States: Yokohama-shi, Kanagawa-ken (JP) AL LT LV MK RO SI • Kohchi, Yasunori Fujisawa-shi, Kanagawa-ken (JP) (30) Priority: 02.06.1997 EP 97108791 • Oikawa, Nobuhiro Yokohama-shi, Kanagawa-ken (JP) (71) Applicant: . Shimma, Nobuo F. HOFFMANN-LA ROCHE AG Chigasaki-shi, Kanagawa-ken (JP) 4070 Basel (CH) . Sudaj Hjtomi Fujisawa-shi, Kanagawa-ken 252 (JP) (72) Inventors: • Hattori, Kazuo Kanagawa-ken (JP) (54) 5'-Deoxy-cytidine derivatives (57) Novel 5'-deoxy-cytidine derivatives repre- group which may be substituted for use in medical ther- sented by the general formula (I) apy, especially tumor therapy. (I) wherein R1 is a hydrogen atom or a group easily hydro- lyzable under physiological conditions; R2 is a hydrogen atom, or -CO-OR4 group [wherein R4 is a saturated or branched CM unsaturated, straight or hydrocarbon group < consisting of one to fifteen carbon atoms, or a group of the formula -(CH2)n-Y (in which Y is cyclohexyl or phe- CO nyl; n is an integer from 0 to 4)]; R3 is a hydrogen atom, r»- bromo, iodo, cyano, a