Diagnostic Proficiency Testing Centre: United Kingdom
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

EXTENDED CARRIER SCREENING Peace of Mind for Planned Pregnancies
Focusing on Personalised Medicine EXTENDED CARRIER SCREENING Peace of Mind for Planned Pregnancies Extended carrier screening is an important tool for prospective parents to help them determine their risk of having a child affected with a heritable disease. In many cases, parents aren’t aware they are carriers and have no family history due to the rarity of some diseases in the general population. What is covered by the screening? Genomics For Life offers a comprehensive Extended Carrier Screening test, providing prospective parents with the information they require when planning their pregnancy. Extended Carrier Screening has been shown to detect carriers who would not have been considered candidates for traditional risk- based screening. With a simple mouth swab collection, we are able to test for over 419 genes associated with inherited diseases, including Fragile X Syndrome, Cystic Fibrosis and Spinal Muscular Atrophy. The assay has been developed in conjunction with clinical molecular geneticists, and includes genes listed in the NIH Genetic Test Registry. For a list of genes and disorders covered, please see the reverse of this brochure. If your gene of interest is not covered on our Extended Carrier Screening panel, please contact our friendly team to assist you in finding a gene test panel that suits your needs. Why have Extended Carrier Screening? Extended Carrier Screening prior to pregnancy enables couples to learn about their reproductive risk and consider a complete range of reproductive options, including whether or not to become pregnant, whether to use advanced reproductive technologies, such as preimplantation genetic diagnosis, or to use donor gametes. -

Amino Acid Disorders
471 Review Article on Inborn Errors of Metabolism Page 1 of 10 Amino acid disorders Ermal Aliu1, Shibani Kanungo2, Georgianne L. Arnold1 1Children’s Hospital of Pittsburgh, University of Pittsburgh School of Medicine, Pittsburgh, PA, USA; 2Western Michigan University Homer Stryker MD School of Medicine, Kalamazoo, MI, USA Contributions: (I) Conception and design: S Kanungo, GL Arnold; (II) Administrative support: S Kanungo; (III) Provision of study materials or patients: None; (IV) Collection and assembly of data: E Aliu, GL Arnold; (V) Data analysis and interpretation: None; (VI) Manuscript writing: All authors; (VII) Final approval of manuscript: All authors. Correspondence to: Georgianne L. Arnold, MD. UPMC Children’s Hospital of Pittsburgh, 4401 Penn Avenue, Suite 1200, Pittsburgh, PA 15224, USA. Email: [email protected]. Abstract: Amino acids serve as key building blocks and as an energy source for cell repair, survival, regeneration and growth. Each amino acid has an amino group, a carboxylic acid, and a unique carbon structure. Human utilize 21 different amino acids; most of these can be synthesized endogenously, but 9 are “essential” in that they must be ingested in the diet. In addition to their role as building blocks of protein, amino acids are key energy source (ketogenic, glucogenic or both), are building blocks of Kreb’s (aka TCA) cycle intermediates and other metabolites, and recycled as needed. A metabolic defect in the metabolism of tyrosine (homogentisic acid oxidase deficiency) historically defined Archibald Garrod as key architect in linking biochemistry, genetics and medicine and creation of the term ‘Inborn Error of Metabolism’ (IEM). The key concept of a single gene defect leading to a single enzyme dysfunction, leading to “intoxication” with a precursor in the metabolic pathway was vital to linking genetics and metabolic disorders and developing screening and treatment approaches as described in other chapters in this issue. -

Prolidase Deficiency
Prolidase deficiency Author: Professor Jaak Jaeken1 Creation Date: August 2001 Update: January 2004 Scientific Editor: Professor Jean-Marie Saudubray 1Center for Metabolic Disease, University Hospital Gasthuisberg, Herestraat 49, B-3000 Leuven, Belgium. [email protected] Abstract Keywords Disease name Excluded diseases Diagnostic criterium Differential diagnosis Prevalence Clinical description Management Etiology Diagnostic methods Genetic counseling Antenatal diagnosis Unresolved questions References Abstract Prolidase deficiency is a very rare autosomal recessive disease. Prevalence remains unknown, some 40 cases have been reported. It is characterized by mild to severe skin lesions particularly on the face, palms, lower legs and soles, besides variable other features. Patients excrete massive amounts of iminodipeptides in urine due to a deficiency of the exopeptidase prolidase. The activity of this enzyme can be measured in hemolysates, leukocytes or fibroblasts. Mutations in the PEPD gene on chromosome 19p13.2 have been observed. The skin ulcers respond partially to local treatment. Keywords prolidase, iminodipeptiduria Disease name Clinical description Prolidase deficiency First symptoms start between birth and young adult age. Most characteristic are the skin Excluded diseases lesions ranging from mild to severe, including Lathyrism recalcitrant ulcerations. They are mostly located on the face, the palms, the lower legs and the Diagnostic criterium soles. Other features are facial dysmorphy, Prolidase deficiency psychomotor retardation and recurrent infections. Differential diagnosis Systemic lupus erythematosus Management This is limited to treatment of the skin ulcers with Prevalence oral ascorbate, manganese (cofactor of Unknown; some 40 individuals have been prolidase), inhibitors of collagenase, and local reported. Jaeken J; Prolidase deficiency. Orphanet encyclopedia, January 2004. http://www.orpha.net/data/patho/GB/uk-prolidase.pdf 1 applications of L-proline-and glycine-containing References ointments. -

Diseases Catalogue
Diseases catalogue AA Disorders of amino acid metabolism OMIM Group of disorders affecting genes that codify proteins involved in the catabolism of amino acids or in the functional maintenance of the different coenzymes. AA Alkaptonuria: homogentisate dioxygenase deficiency 203500 AA Phenylketonuria: phenylalanine hydroxylase (PAH) 261600 AA Defects of tetrahydrobiopterine (BH 4) metabolism: AA 6-Piruvoyl-tetrahydropterin synthase deficiency (PTS) 261640 AA Dihydropteridine reductase deficiency (DHPR) 261630 AA Pterin-carbinolamine dehydratase 126090 AA GTP cyclohydrolase I deficiency (GCH1) (autosomal recessive) 233910 AA GTP cyclohydrolase I deficiency (GCH1) (autosomal dominant): Segawa syndrome 600225 AA Sepiapterin reductase deficiency (SPR) 182125 AA Defects of sulfur amino acid metabolism: AA N(5,10)-methylene-tetrahydrofolate reductase deficiency (MTHFR) 236250 AA Homocystinuria due to cystathionine beta-synthase deficiency (CBS) 236200 AA Methionine adenosyltransferase deficiency 250850 AA Methionine synthase deficiency (MTR, cblG) 250940 AA Methionine synthase reductase deficiency; (MTRR, CblE) 236270 AA Sulfite oxidase deficiency 272300 AA Molybdenum cofactor deficiency: combined deficiency of sulfite oxidase and xanthine oxidase 252150 AA S-adenosylhomocysteine hydrolase deficiency 180960 AA Cystathioninuria 219500 AA Hyperhomocysteinemia 603174 AA Defects of gamma-glutathione cycle: glutathione synthetase deficiency (5-oxo-prolinuria) 266130 AA Defects of histidine metabolism: Histidinemia 235800 AA Defects of lysine and -
Biochemical Basis of Prolidase Deficiency
Biochemical Basis of Prolidase Deficiency Polypeptide and RNA Phenotypes and the Relation to Clinical Phenotypes Fumio Endo,* Akito Tanoue,* Akito Kitano,* Jiro Arata,t David M. Danks,9 Charles M. Lapiere,1 Yoshihiro Sei,1 Sybe K. Wadman,** and Ichiro Matsuda* *Department ofPediatrics, Kumamoto University Medical School, Kumamoto, Japan; tDepartment ofDermatology, Okayama University Medical School, Okayama, Japan; §The Murdoch Institute, Royal Children's Hospital, Victoria, Australia; IlDepartment of Dermatology, University ofLiege, Liege, Belgium; 'Department ofDermatology, Kanazawa Medical University, Uchinada, Japan; and ** Wilhelmina Children's Hospital, Utrecht, The Netherlands Abstract sis of di- and tripeptides with carboxy-terminal proline and Cultured skin fibroblasts or lymphoblastoid cells from eight seems to play an important role in the recycling of proline. As patients with clinical symptoms of prolidase deficiency were a consequence of the deficiency of prolidase, massive imido- analyzed in terms of enzyme activity, presence of material peptiduria and relatively lowered levels of plasma proline are antibodies, biosynthesis of the poly- present in the patients (1). The precise mechanisms involved crossreacting with specific mental retardation, and peptide, and mRNA corresponding to the enzyme. There are at in the development of skin lesions, least two enzymes that hydrolyze imidodipeptides in these cells other symptoms including splenomegaly, osteoporosis, and and these two enzymes could be separated by an immunochem- frequent infections have not been completely elucidated. ical procedure. The specific assay for prolidase showed that More than 20 cases of prolidase deficiency have been re- the enzyme activity was virtually absent in six cell strains and ported, and some of the patients had no clinical symptoms of was markedly reduced in two (< 3% of controls). -

Prolidase Deficiency: a New Genetic Cause of Combined Pulmonary Fibrosis and Emphysema Syndrome in the Adult
Early View Research letter Prolidase deficiency: a new genetic cause of combined pulmonary fibrosis and emphysema syndrome in the adult Vincent Cottin, Mouhamad Nasser, Julie Traclet, Lara Chalabreysse, Anne-Sophie Lèbre, Salim Si- Mohamed, François Philit, Françoise Thivolet-Béjui Please cite this article as: Cottin V, Nasser M, Traclet J, Chalabreysse L, et al. Prolidase deficiency: a new genetic cause of combined pulmonary fibrosis and emphysema syndrome in the adult. Eur Respir J 2020; in press (https://doi.org/10.1183/13993003.01952-2019). This manuscript has recently been accepted for publication in the European Respiratory Journal. It is published here in its accepted form prior to copyediting and typesetting by our production team. After these production processes are complete and the authors have approved the resulting proofs, the article will move to the latest issue of the ERJ online. Copyright ©ERS 2020 Prolidase deficiency: a new genetic cause of combined pulmonary fibrosis and emphysema syndrome in the adult Letter to the editor Authors Vincent Cottin 1a, Mouhamad Nasser 1, Julie Traclet 1, Lara Chalabreysse 2, Anne-Sophie Lèbre3, Salim Si-Mohamed 4, François Philit 5, Françoise Thivolet-Béjui 2 ORCID : V. Cottin : 0000-0002-5591-0955; M. Nasser : 0000-0001-8373-8032; A.S. Lebre : 0000-0002-7519-9822 Affiliations 1 National Coordinating Reference Centre for Rare Pulmonary Diseases, Louis Pradel Hospital, Hospices Civils de Lyon, UMR 754, Claude Bernard University Lyon 1, member of OrphaLung, RespiFil, and ERN-LUNG, Lyon, -

Diagnose a Broad Range of Metabolic Disorders with a Single Test, Global
PEDIATRIC Assessing or diagnosing a metabolic disorder commonly requires several tests. Global Metabolomic Assisted Pathway Screen, commonly known as Global MAPS, is a unifying test GLOBAL MAPS™ for analyzing hundreds of metabolites to identify changes Global Metabolomic or irregularities in biochemical pathways. Let Global MAPS Assisted Pathway Screen guide you to an answer. Diagnose a broad range of metabolic disorders with a single test, Global MAPS Global MAPS is a large scale, semi-quantitative metabolomic profiling screen that analyzes disruptions in both individual analytes and pathways related to biochemical abnormalities. Using state-of-the-art technologies, Global Metabolomic Assisted Pathway Screen (Global MAPS) provides small molecule metabolic profiling to identify >700 metabolites in human plasma, urine, or cerebrospinal fluid. Global MAPS identifies inborn errors of metabolism (IEMs) that would ordinarily require many different tests. This test defines biochemical pathway errors not currently detected by routine clinical or genetic testing. IEMs are inherited metabolic disorders that prevent the body from converting one chemical compound to another or from transporting a compound in or out of a cell. NORMAL PROCESS METABOLIC ERROR These processes are necessary for essentially all bodily functions. Most IEMs are caused by defects in the enzymes that help process nutrients, which result in an accumulation of toxic substances or a deficiency of substances needed for normal body function. Making a swift, accurate diagnosis -
Blueprint Genetics Comprehensive Immune and Cytopenia Panel
Comprehensive Immune and Cytopenia Panel Test code: IM0901 Is a 642 gene panel that includes assessment of non-coding variants. Is ideal for patients with a clinical suspicion of an inborn error of immunity, such as, Primary Immunodeficiency, Bone Marrow Failure Syndrome, Dyskeratosis Congenita, Neutropenia, Thrombocytopenia, Hemophagocytic Lymphohistiocytosis, Autoinflammatory Disorders, Complement System Disorder, Leukemia, or Chronic Granulomatous Disease. This panel includes most genes from Primary Immunodeficiency, Severe Combined Immunodeficiency, Complement System Disorder, Bone Marrow Failure Syndrome, Hemophagocytic Lymphohistiocytosis, Congenital Neutropenia, Thrombocytopenia, Congenital Diarrhea, Chronic Granulomatous Disease, Diamond-Blackfan Anemia, Fanconi Anemia, Dyskeratosis Congenita, Autoinflammatory Syndrome, and Hereditary Leukemia Panels as well as many other genes associated with inborn errors of immunity. Please note that unlike our other panels, this panel is on our Whole Exome Sequencing platform and cannot be customized. Pricing may vary from our regular panel pricing. About immunodeficiency and cytopenia disorders There is an enormous amount of phenotypic overlap between immunological and hematological disorders, which makes it challenging to know which of these two systems is not functioning properly. Knowing the underlying genetic cause of a person’s clinical diagnosis, especially immunodeficiency, bone marrow failure, neutropenia, thrombocytopenia, autoinflammatory disease, or bone marrow failure can sometime -
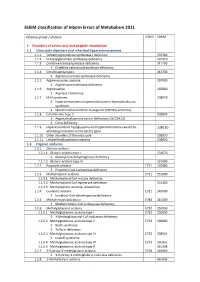
SSIEM Classification of Inborn Errors of Metabolism 2011
SSIEM classification of Inborn Errors of Metabolism 2011 Disease group / disease ICD10 OMIM 1. Disorders of amino acid and peptide metabolism 1.1. Urea cycle disorders and inherited hyperammonaemias 1.1.1. Carbamoylphosphate synthetase I deficiency 237300 1.1.2. N-Acetylglutamate synthetase deficiency 237310 1.1.3. Ornithine transcarbamylase deficiency 311250 S Ornithine carbamoyltransferase deficiency 1.1.4. Citrullinaemia type1 215700 S Argininosuccinate synthetase deficiency 1.1.5. Argininosuccinic aciduria 207900 S Argininosuccinate lyase deficiency 1.1.6. Argininaemia 207800 S Arginase I deficiency 1.1.7. HHH syndrome 238970 S Hyperammonaemia-hyperornithinaemia-homocitrullinuria syndrome S Mitochondrial ornithine transporter (ORNT1) deficiency 1.1.8. Citrullinemia Type 2 603859 S Aspartate glutamate carrier deficiency ( SLC25A13) S Citrin deficiency 1.1.9. Hyperinsulinemic hypoglycemia and hyperammonemia caused by 138130 activating mutations in the GLUD1 gene 1.1.10. Other disorders of the urea cycle 238970 1.1.11. Unspecified hyperammonaemia 238970 1.2. Organic acidurias 1.2.1. Glutaric aciduria 1.2.1.1. Glutaric aciduria type I 231670 S Glutaryl-CoA dehydrogenase deficiency 1.2.1.2. Glutaric aciduria type III 231690 1.2.2. Propionic aciduria E711 232000 S Propionyl-CoA-Carboxylase deficiency 1.2.3. Methylmalonic aciduria E711 251000 1.2.3.1. Methylmalonyl-CoA mutase deficiency 1.2.3.2. Methylmalonyl-CoA epimerase deficiency 251120 1.2.3.3. Methylmalonic aciduria, unspecified 1.2.4. Isovaleric aciduria E711 243500 S Isovaleryl-CoA dehydrogenase deficiency 1.2.5. Methylcrotonylglycinuria E744 210200 S Methylcrotonyl-CoA carboxylase deficiency 1.2.6. Methylglutaconic aciduria E712 250950 1.2.6.1. Methylglutaconic aciduria type I E712 250950 S 3-Methylglutaconyl-CoA hydratase deficiency 1.2.6.2. -

Blueprint Genetics Organic Acidemia/Aciduria &Amp
Organic Acidemia/Aciduria & Cobalamin Deficiency Panel Test code: ME0901 Is a 54 gene panel that includes assessment of non-coding variants. Is ideal for patients with a clinical suspicion of cobalamin deficiency, homocystinuria, maple syrup urine disease, methylmalonic acidemia, organic acidemia/aciduria or propionic acidemia. The genes on this panel are included in the Comprehensive Metabolism Panel. About Organic Acidemia/Aciduria & Cobalamin Deficiency Organic acidemia and aciduria refer to many disorders, where non-amino organic acids are excreted in urine. This is usually a result of deficient enzyme activity in amino acid catabolism. The clinical presentation of organic acidemia in young children includes neurologic symptoms, poor feeding and lethargy progressing to coma. Older persons with this disorder often also have neurological signs, recurrent ketoacidosis and loss of intellectual function. The symptoms result from the damaging accumulation of precursors of the defective pathway. The combined prevalence of organic acidurias is estimated at 1:1,000 newborns. Cobalamin, also known as vitamin B12, has cobalt in its structure. Humans are not able to synthesize B12. It must therefore be obtained from a food of animal origin (the only natural source of cobalamin in the human diet). Intracellular cobalamin deficiencies can be subgrouped based on the cellular complementation groups and defective genes. Mutations in genes MMAA, MMAB and MMADHC cause deficient synthesis of the coenzyme adenosylcobalamin (AdoCbl), while mutations in genes MMADHC, MTRR and MTR cause defective methylcobalamin (MeCbl) synthesis. Mutation in genes MMACHC, MMADHC, LMBRD1 and ABCD4 result in combined AdoCbl and MeCbl deficiency. Mutations in MMACHC explain approximately 80% of the cases with intracellular cobalamin deficiency, followed by MMADHC (<5%), TRR (<5%), LMBRD1 (<5%), MTR (<5%) and ABCD4 (<1%). -

1 a Clinical Approach to Inherited Metabolic Diseases
1 A Clinical Approach to Inherited Metabolic Diseases Jean-Marie Saudubray, Isabelle Desguerre, Frédéric Sedel, Christiane Charpentier Introduction – 5 1.1 Classification of Inborn Errors of Metabolism – 5 1.1.1 Pathophysiology – 5 1.1.2 Clinical Presentation – 6 1.2 Acute Symptoms in the Neonatal Period and Early Infancy (<1 Year) – 6 1.2.1 Clinical Presentation – 6 1.2.2 Metabolic Derangements and Diagnostic Tests – 10 1.3 Later Onset Acute and Recurrent Attacks (Late Infancy and Beyond) – 11 1.3.1 Clinical Presentation – 11 1.3.2 Metabolic Derangements and Diagnostic Tests – 19 1.4 Chronic and Progressive General Symptoms/Signs – 24 1.4.1 Gastrointestinal Symptoms – 24 1.4.2 Muscle Symptoms – 26 1.4.3 Neurological Symptoms – 26 1.4.4 Specific Associated Neurological Abnormalities – 33 1.5 Specific Organ Symptoms – 39 1.5.1 Cardiology – 39 1.5.2 Dermatology – 39 1.5.3 Dysmorphism – 41 1.5.4 Endocrinology – 41 1.5.5 Gastroenterology – 42 1.5.6 Hematology – 42 1.5.7 Hepatology – 43 1.5.8 Immune System – 44 1.5.9 Myology – 44 1.5.10 Nephrology – 45 1.5.11 Neurology – 45 1.5.12 Ophthalmology – 45 1.5.13 Osteology – 46 1.5.14 Pneumology – 46 1.5.15 Psychiatry – 47 1.5.16 Rheumatology – 47 1.5.17 Stomatology – 47 1.5.18 Vascular Symptoms – 47 References – 47 5 1 1.1 · Classification of Inborn Errors of Metabolism 1.1 Classification of Inborn Errors Introduction of Metabolism Inborn errors of metabolism (IEM) are individually rare, but collectively numerous. -

Onset Lupus Frequently Detects Single Gene Etiologies
Tirosh et al. Pediatric Rheumatology (2019) 17:52 https://doi.org/10.1186/s12969-019-0349-y RESEARCH ARTICLE Open Access Whole exome sequencing in childhood- onset lupus frequently detects single gene etiologies Irit Tirosh1,2,8, Shiri Spielman2,8, Ortal Barel10, Reut Ram1, Tali Stauber3,8, Gideon Paret4,8, Marina Rubinsthein4,8, Itai M. Pessach4,8, Maya Gerstein1,8, Yair Anikster5,8, Rachel Shukrun1,8, Adi Dagan1,8, Katerina Adler10, Ben Pode-Shakked1,5,8, Alexander Volkov6,8, Marina Perelman6,8, Shoshana Greenberger7,8, Raz Somech3,8, Einat Lahav3,8,11, Amar J. Majmundar9, Shai Padeh1,8, Friedhelm Hildebrandt9 and Asaf Vivante1,8,11* Abstract Background: Systemic lupus erythematosus (SLE) comprise a diverse range of clinical manifestations. To date, more than 30 single gene causes of lupus/lupus like syndromes in humans have been identified. In the clinical setting, identifying the underlying molecular diagnosis is challenging due to phenotypic and genetic heterogeneity. Methods: We employed whole exome sequencing (WES) in patients presenting with childhood-onset lupus with severe and/or atypical presentations to identify cases that are explained by a single-gene (monogenic) cause. Results: From January 2015 to June 2018 15 new cases of childhood-onset SLE were diagnosed in Edmond and Lily Safra Children’s Hospital. By WES we identified causative mutations in four subjects in five different genes: C1QC, SLC7A7, MAN2B1, PTEN and STAT1. No molecular diagnoses were established on clinical grounds prior to genetic testing. Conclusions: We identified a significant fraction of monogenic SLE etiologies using WES and confirm the genetic locus heterogeneity in childhood-onset lupus.