INTERLABORATORY COMPARISON on Pops in FOOD 2020: the Twenty-First Round of an International Study
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Solceller Og Grundvandsbeskyttelse
Solceller og grundvandsbeskyttelse Jan Vedde European Energy A/S Agenda Hvordan kan solcelleanlæg og grundvandsbeskyttelse understøtte hinanden? • Hvem er European Energy og hvad laver vi? • Hvorfor alle disse solcelleanlæg på markerne? • Hvordan får man tilladelse til at etablere en solcellepark? • Konkrete aktiviteter omkring BNBO • Specifikke miljømæssige temaer: • Montagestrukturer (faste stativer og tracker) • PV moduler (inkl. glascoating) • Elektriske komponenter (invertere & kabler) • Transformatorer • D&V inklusiv rengøring • Flora, fauna og biodiversitet • Afrunding European Energy – Jan Vedde - [email protected] PAGE 2 European Energy is built on four pillars Innovation and Solar power Onshore wind Offshore wind Energy storage Active in Europe, Americas, Active in Europe and Brazil Active in Europe Active in Europe and Australia 185 employees in European Energy PAGE 3 Our business model Asset Development Construction Divestment Management • Potential wind & solar • Select optimal • Sale and handover of • Protect returns for site assessment technology and park wind and solar farms to investors and partners by • Project risk assessment layouts long-term investors optimising production output • Environmental studies • Oversee every • Internal M&A team with construction phase from 11 employees • Identifying repowering • Secure land & building opportunities within the permits groundworks to grid • Internal legal department connection with 10 employees operational portfolio • Approx. 60 people work • AM team of 14 within development (both • Approx. 30 employees employees external and internal) work within construction Project financing Production and sale of electricity • For each project European Energy seeks the optimal capital partner. • Independent production of electricity • In 2019, European Energy renewed the Bond loan with EUR 200m • Sale of electricity to the grid and raised or refinanced project loans of more than EUR 85m. -

Indsatsplan for Grundvandsbeskyttelse I Slagelse Nordøst Indsatsområde
HØRINGSFORSLAG • FEBRUAR 2006 Indsatsplan for grundvandsbeskyttelse i Slagelse nordøst indsatsområde KongstedKongstedHolmeHolme OOrevadOOrevadrevarevarevadd KongstedKongstedHolmeHolme FrederiksbergFrederiksberg KongstedKongsted ÅgerupÅgerup TystrupTystrup SobankeSobanke VerupVerup AAssentorssentorssentorp SobankeSobanke NyNy TågerupTågerup LårupLårup LundenLunden vedved TissøTissø NyNy TågerupTågerup NiløseNiløse ReerslevReerslev HoltehHoltehu OugtvedOugtved TågerupTågerup OreboOrebo LambækLambæk SkuerupSkuerup HallenslevHallenslevSæbySæby HedeborydeHedeboryde SæbySæby HestehaveHestehave HedeborydeHedeboryde BrandstrupBrandstrup StenlillStenlillee RudsRuds VedbyVedby BrandstrupBrandstrup GørlevGørlev UlstrupUlstrup HjorthøjHjorthøj GårdeGårde SkellebjergSkellebjerg TjørntvedTjørntved HjorthøjHjorthøj GårdeGårde SkellebjergSkellebjerg VeddeVedde Tude Å kortlægningsområde DianalundDianalund DianalundDianalund SaltofteSaltofte SandhagenSandhagen VindeVinde HelsingeHelsinge TorpeTorpe HerslevHerslev HalshuseHalshuse SobjergSobjerg HalshuseHalshuse RyeRye KulbyKulby KammergaveKammergave HuseHuse ReersøReersø VejrbækVejrbæk ToppeneToppene VejrbækVejrbæk ToppeneToppene TåderupTåderup TersløseTersløse DalbyDalby KirkeKirke HelsingeHelsinge FlaskehuseFlaskehuse TjørnelundeTjørnelunde TåderupTåderup MunkeMunke BjergbyBjergby DøjringeDøjringe HusHusee VrangeVrange HuseHuse SkjoldmøsSkjoldmøs HuseHuse SkætholmSkætholm MunkeMunke BjergbyBjergby HøngHøng TangenTangen FastemoseFastemose HundekrogHundekrog StokkehuseStokkehuse LodbjerggårdLodbjerggård -

Roskilde · 44 21 64 30 · 8 4 10 2 6 2 5 Søren Frid 1
1 9 Åbningstider: Mandag - Lukket Tirsdag - 10-18 Onsdag - 10-18 Torsdag - 10-18 Fredag - 10-18 Lørdag - 9-15 3 Søndag - Lukket 7 Skomagergade 1 · Roskilde · 44 21 64 30 · www.caseus.dk 8 4 10 2 6 2 5 Søren Frid 1 Optique 60 BUTIKKER • 12 SPISESTEDER DAGLIGVARER • BIOGRAF • FITNESS HVERDAGE KL. 10-19 Stændertorvet 6 WEEKEND KL. 10-17 4000 Roskilde ROSTORV.DK Tlf. 4634 1234 3 XXXX_annonce_70x96_bykort.indd 1 21/11/201910 16.38 ZLEEP HOTEL ROSKILDE [ ALSO KNOWN AS HOTEL PRINDSEN ] PREVIOUSLY VISITED BY H.C. ANDERSEN IN THE HEART OF ROSKILDE YOU’LL FIND THIS PEARL OF A HOTEL WITH A UNIQUE HISTORY WHERE MODERN NORDIC DESIGN MEETS OLD BEAUTIFUL ROKOKO INTERIOR Vestre Kirkevej 12, Himmelev • 4000 Roskilde Tlf. 46 32 31 31 • [email protected] SEE OUR MANY OTHER HOTELS AT WWW.ZLEEP.COM 9 4 RESTAURANT UPSTAIRS Skomagergade 4 1. sal / 1st floor ROSKILDE 4000 Roskilde Tlf. 46 35 19 00 ÅBEN / OPEN MAN-LØR KL 11-22 BRUNCH / FROKOST KL 11-16 SØN LUKKET / CLOSED AFTENMENU KL 17.30-21 Roskilde 5 6 7 8 Algade 24 SørenSøren Frid Frid Anytime Optique Ure & Smykker Optique Watches and Jewellery Stændertorvet 6 4000 RoskildeStændertorvet 6 Tlf. 4634 12344000 Roskilde Åbningstider: Tlf. 4634 1234 Algade 21 - 4000 Roskilde - 46365373 Mandag - Lukket SUPERMARKED • Algade 51 • tlf. +45 46 34 03 10 • www.kvickly.dk www.anytimeureogsmykker.dk Tirsdag - 10-18 Onsdag - 10-18 BYKORTTorsdag - 10-18 / MAP / STADTKARTE Fredag - 10-18 Lørdag - 9-15 OMRÅDEKORTSøndag - Lukket / AREA MAP / KARTE Skomagergade 1 · Roskilde · 44 21 64 30 · www.caseus.dk Nellerød Rørt -

Sorø Kommune Detailhandelsanalyse Sorø Kommuneplan 2009
Sorø Kommune Detailhandelsanalyse Sorø Kommuneplan 2009-2020 Maj 2009 Version 060509 Sorø Kommune Side i Detailhandelsanalyse – Sorø Kommuneplan 2009 Maj 2009 INDHOLD 1. INDLEDNING OG BAGGRUND............................................................1 1.1 Indledning........................................................................................1 1.2 Ændrede detailhandelsregler i planloven ........................................2 1.3 Centerstrukturen i Sorø Kommune ................................................. 2 1.3.1 Bymidter ..............................................................................3 1.3.2 Lokalcentre .........................................................................3 1.3.3 Butikker til pladskrævende varegrupper............................. 4 1.3.4 Enkeltstående butikker ........................................................4 1.3.5 Salg af egne produkter ........................................................4 1.4 Rapportens opbygning.....................................................................4 2. BYRÅDETS MÅL ....................................................................................5 3. REDEGØRELSE.......................................................................................6 3.1 Generelle udviklingsperspektiver ....................................................6 3.1.1 Internethandel og postordrevirksomheder ..........................6 3.1.2 Ændring af lukkeloven .......................................................6 3.2 Eksisterende datagrundlag og butiksstruktur...................................7 -
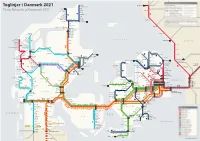
Toglinjer I Danmark 2021
Toglinjer i Danmark 2021 Skagen Göteborg C Frederikshavnsvej Hirtshals Hulsig Train Network of Denmark 2021 Lilleheden Bunken Mölndal Emmersbæk Horne Ålbaek N Tornby Napstjært Ø Vidstrup N Jerup Vellingshøj Herregårdsparken Rimmen Teglgårdsvej Hjørring Tolne Strandby Kungsbacka Kvægtorvet Sindal Kvissel Frederikshavn Vrå Brønderslev Varberg Aalborg Lufthavn Lindholm Aalborg Vestby Aalborg Skalborg Thisted Falkenberg Sjørring Svenstrup Støvring Snedsted Skørping Hørdum Thyborøn Bedsted Thy KATTEGAT RA Arden Halmstad C Thyborøn Kirke Hurup Thy SVERIGE Sprogøvej Ydby Rønland Lyngs Hobro Laholm Harboøre Hvidbjerg M IC Vrist Uglev Victoria Street Station Oddesund Nord Skive Stoholm RA Lyn Strande Humlum Højslev Båstad Nejrup Balleby Lemvig Randers Struer Vinderup Viborg Gilleleje Klinkby Armose Hornbæk Rødkærsbro Bjerringbro Langå Tisvildeleje Bonnet Hjerm L Ängelholm Ramme Grenaa Dronning- Ålsgårde Fåre Ulstrup L2 Ryomgård mølle L Sinkbæk Holstebro Lisbjergskolen Lystrup Vejby Grønnehave Græsted Bækmarksbro Hadsten Hornslet Universitets- Skødstrup Helsinge Helsingborg C Amstrup Bur hospitalet Hundested Aulum Hinnerup Torsøvej Hundested Havn Vibehus Kagerup Helsingør Vemb Nyled Melby Snekkersten Ulfborg Vildbjerg Herning Hammerum Bording Silkeborg L1 Slotspavillonen L M Viby J Højby Nykøbing Sj Espergærde Stockholm Tim Gødstrup Svejbæk Fredensborg Kalmar Engesvang Sommerland Sj L Humlebæk Karlskrona Birk Ikast Laven Østbanetorvet Frederiksværk Ø Hee Herning Messecenter Center- Hørning Kvistgård ↗ RA Nr. Asmindrup Lille Kregme Hillerød Nivå -

Baggrundsmateriale Movia
Notat Til: Sagsnummer ThecaSag-115674 Movit-3007819 Region Sjælland Sagsbehandler TFR Kopi til: Direkte +45 36 13 16 00 Fax - [email protected] CVR nr: 29 89 65 69 EAN nr: 5798000016798 29. januar 2015 Tilpasningsbehov i Region Sjælland D Forvaltningen i Region Sjælland har bedt Movias administration bidrage med et katalog, der beskriver muligheder for og konsekvenser ved en reduktion i tilskudsbehovet på 30 mio. kr. til den kollektive trafik, til brug for Regionens videre arbejde med budgettet for Regional Ud- vikling for 2016 og frem. Med Movias Trafikplan 2013 sættes der nye mål. Målene er besluttet politisk efter høring i kommuner og regioner. I Region Sjællands område er målet 3,3 mio. flere påstigere frem mod 2020, svarende til en fremgang på omkring 10 procent. Samtidig betyder den ny lovgivning for trafikselskaber, at regionen fra 2016 overtager ansva- ret for en række buslinjer fra kommunerne, og dermed kommer til at spille en endnu større rolle ift. opfyldelsen af målene i Movias Trafikplan 2013. I Budget 2015 har Region Sjælland udgifter på lige knap 100 mio. kr. til bustrafik. En budget- reduktion i størrelsen 30 mio. kr. vil derfor i et scenarie, hvor beløbet findes ved driftsredukti- oner i den kollektive trafik, naturligt have meget negative konsekvenser ift. de øvrige regio- nalpolitiske målsætninger, som Region Sjælland arbejder med Forvaltningen har anmodet om en opstilling af løsningsmuligheder, der anviser forskellige veje til at budgetreduktion kan indfries, uanset om Tølløsebanen forudsættes nedlagt og er- stattet af en busløsning, eller om banen fortsætter i drift. I Der er derfor lavet en opstilling af mulige løsninger, hvor de enkelte muligheder er opstillet som separate ”pakker”, som kan medtages eller udelades i de videre politiske drøftelser uafhængigt af hinanden. -

Icelandic Volcanic Ash from the Late-Glacial Open-Air Archaeological Site of Ahrenshöft LA 58 D, North Germany
Journal of Archaeological Science 39 (2012) 708e716 Contents lists available at SciVerse ScienceDirect Journal of Archaeological Science journal homepage: http://www.elsevier.com/locate/jas Icelandic volcanic ash from the Late-glacial open-air archaeological site of Ahrenshöft LA 58 D, North Germany R.A. Housley a,*, C.S. Lane b, V.L. Cullen b, M-J. Weber c, F. Riede d, C.S. Gamble e,F.Brockb a Department of Geography, Royal Holloway, University of London, Egham Hill, Egham TW20 0EX, UK b Research Laboratory for Archaeology and History of Art, University of Oxford, Dyson Perrins Building, South Parks Road, Oxford OX1 3QY, UK c Zentrum für Baltische und Skandinavische Archäologie, Stiftung Schleswig-Holsteinische Landesmuseen, Schloss Gottorf, D-24837 Schleswig, Germany d Afdeling for Forhistorisk Arkæologi, Institut for Antropologi, Arkæologi og Lingvistik, University of Aarhus, Moesgård Allé 20, DK-8270 Højbjerg, Denmark e Faculty of Humanities (Archaeology), Building 65A, Avenue Campus, University of Southampton, Southampton SO17 1BF, UK article info abstract Article history: Cryptotephra of Icelandic origin from the open-air archaeological site of Ahrenshöft LA 58 D Received 28 March 2011 (Kr. Nordfriesland, Schleswig-Holstein), northern Germany overlies a Late-glacial Havelte lithic assem- Received in revised form blage, hitherto dated by 14C and biostratigraphy to the earliest part of the Late-glacial interstadial (GI-1e 31 October 2011 to GI-1c ). Peaks in ash shards are observed in two profiles. Major and minor element geochemistry Accepted 1 November 2011 3 indicates volcanic ash originating in the Katla system. Precise correlation to previously described tephra is uncertain due to overlapping chemical characteristics. -

DK19786/2012 Køn: H Race: Stabyhoun Født: 22/10-2012 Farve: Sort M Hvide Aftegn Indavlsgrad: 0% (Beregnet På 3 Generationer) HD: a AD: 0
Dansk Kennel Klubs hundedatabase på internettet Dansk Kennel Klub · Parkvej 1 · 2680 Solrød Strand · Denmark Dette er en udskrift fra DKK's hundedatabase på DKK's officielle hjemmeside www.hundeweb.dk. Denne udskrift erstatter ikke hundens orginale stamtavle. This is a printout from DKK's dog database on DKK's webpage www.hundeweb.dk. This printout does not replace the dogs original pedigree Hundens navn: Herkules Regnr: DK19786/2012 Køn: H Race: Stabyhoun Født: 22/10-2012 Farve: Sort M Hvide Aftegn Indavlsgrad: 0% (Beregnet på 3 generationer) HD: A AD: 0 Hundens far: Spyke Fan 'e Blommetún NHSB 2650072 HD:A Hundens mor: Abildkær's Anuka Van Benji DK09180/2007 HD:A F:S/HVA Bedsteforældre Oldeforældre Tip-Oldeforældre Midas Buck NHSB 1056332 HD:C NHSB 2213643 HD:C Douktje Femke Fan 't Romme Pjild Feike Fan De Fryske Strúners NHSB 1853330 HD:B NHSB 2408983 Aelderd-Bjintse V.D. Kraay Heide Heidi NHSB 1955476 HD:B HD:B NHSB 2272726 HD:B Linke Fan 't Sudermiem Fars aner Fars NHSB 2073676 HD:C NLCH WW02 Djoeke V.D. Zodde Vedde Elvis V.D. Vronergeest NHSB 1987032 HD:B NHSB 2202115 HD:C Rapke Cleopatra V.D. Vronergeest Jikkejolien NHSB 1873727 HD:C NHSB 2331205 Ivar V.'t Hooge Holt Tjamke NHSB 1940160 HD:C HD:A NHSB 2142279 HD:B Reja Fan It Drentse Hiem NHSB 1973240 HD:B NLCH WW95 Frids Fan'e Ikeloane Bluf NHSB 1786282 HD:C NHSB 2032653 HD:A Hiske Femke Fan't Paradijske Stabygårds Benji Von Bluf NHSB 1780247 HD:B 00822/2004 Otte Ludo Bart Fan It Heidehiem Inez V.D. -

8 Alternativer
8 Alternativer I en VVM-proces er der undervejs i fastlæg- gelsen af projektet mange alternativer i spil – f.eks. i form af forskellige linieføringer, teknisk udformning af anlægskomponenter m.v. Her er en beskrivelse af de udbygnings- forslag samt alternativer til linieføringer og placering af kompressorstationer, som har været i spil i VVM-processen. 136 137 I dette kapitel præsenteres alternativerne til de udbygningsforslag der er beskrevet og økonomiske årsager. Nedenfor præsenteres de fravalgte alternativer og de specifikke konsekvensvurderet i kapitel 6. Endvidere præsenteres de alternativer til linieføringer og årsager til at de er fravalgt. placeringer af kompressorstationer som er blevet fravalgt. Vejle Amt Nul-alternativet I Vejle Amt har en række alternative linieføringer været undersøgt. Særligt ved Elbodalen Nul-alternativet beskriver den situation at der ikke leveres yderligere gas på den jyske er flere forløb undersøgt, men også syd for Egtved, ved Vester Nebel og Viuf og ved vestkyst. De overordnede konsekvenser af nul-alternativet til BalticPipe og udvidelser på Erritsø er en del alternativer valgt fra i undersøgelsesforløbet. tværs af Danmark vil være : • at Syd Arne terminalen i Nybro ikke skal udvides • at der ikke skal foretages anlægsudvidelser fra Egtved til den sjællandske østkyst • at der ikke skal etableres en søledningsforbindelse fra Danmark til Polen • at Danmark afskærer sig fra muligheden for at opnå en strategisk placering på det nordeuropæiske gasmarked • at Danmark mister muligheden for på længere sigt at importere russisk gas direkte gennem en polsk-dansk forbindelse når de danske gasleverancer efter 2010 forventes at falde. Nul-alternativet vil betyde at samtlige konsekvenser af anlægsarbejderne for natur og miljø undgås. -

Indsatsplan for Grundvandsbeskyttelse Sorø-Stenlille Og Fjenneslev
INDSATSPLAN FOR GRUNDVANDSBESKYTTELSEN I SORØ-STENLILLE OG FJENNESLEV OMRÅDET SORØ KOMMUNE 2018 INDSATSPLAN FOR GRUNDVANDSBESKYTTELSEN I SORØ-STENLILLE OG FJENNESLEV OMRÅDET Dato 2018-12-05 Udarbejdet af TEO, NLR. Rambøll og CHHO for Sorø Kommune Indsatsplan for grundvandsbeskyttelse for Sorø-Stenlille og Fjenneslev området i Sorø Kommune INDHOLD 1. RESUME 4 2. INDLEDNING 6 3. LOVGIVNING 7 4. RESUME AF GRUNDVANDSKORTLÆGNING 8 4.1 Vandforsyningsstruktur 9 4.2 Arealanvendelse og forureningskilder 9 4.3 Grundvandsmagasiner og dæklag 11 4.4 Grundvandets strømning 14 4.5 Grundvandets kvalitet 17 4.6 Sårbarhed og områdeudpegninger 19 5. GENERELLE INDSATSER 22 5.1 Områder omfattet af indsatsplanen 22 5.2 Boringsnære beskyttelsesområder (BNBO) 23 5.3 Indsatser i forhold til nitrat 24 5.4 Indsatser i forhold til pesticider 26 5.5 Indvindingsstrategi 28 5.6 Grundvandsovervågning 28 5.7 Indsatser i forhold til forurenede lokaliteter 29 5.8 Indsatser i forhold til potentielt grundvandstruende virksomheder 29 5.9 Lokalisering og sløjfning af ubenyttede boringer og brønde 30 5.10 Indsatser i forhold til spildevandsslam 31 5.11 Pesticidoplysningskampagner til private 31 5.12 Indsatser i forhold til råstofområder 32 5.13 Skovrejsning 32 5.14 Indsatser i forhold til nedsivningsanlæg og spildevandsledninger/-kloaker 33 5.15 Indsatser i forhold til jordvarmeanlæg 35 5.16 Indsatser i forhold til varmeindvindings- og grundvandskøleanlæg 36 6. SPECIFIKKE INDSATSER 37 6.1 Bjernede Vandværk 37 6.2 Dianalund Vandværk 40 6.3 Døjringe Vandværk, Dybendal kildeplads 48 6.4 Døjringe Vandværk, Døjringe kildeplads 51 6.5 Fjenneslev Vandværk 54 6.6 Frederiksberg Vandværk 58 6.7 Munke Bjergby Vandværk 67 6.8 Nyrup Vandværk 71 6.9 Sorø Vandværk 75 6.10 Stenlille Vandværk 89 6.11 Vedde Vandværk 94 7. -

Slagelse-Kommunekort.Pdf
Liseleje 327 Gniben Lindeb Gnib 327 561 enve j jerg yve j lb ve Me j Gn Sjællands ibe Odde Melby St. 56 nvej Dyssekilde St. 1 Odd enve j j Odden e Torup env Kikhavn 561 Fæ Færgehavn dd r O 5 gevej 555 55 5 e Østerbjerg 561 555 61 n Yderby Ve 561 555 stre 56 Hav Hanehoved 1 nev Korshage ej 326 Lille Ka Havnebyen rlsmindeve Hundested Havn j j 561 555 edvej 323266 anehov Overby Ullerup Amager Huse H Klintebjerg Pe 561 555 der Stadionve 326 555 Oddenvej Dybesø Hundested St. Olsensv Am n 5 Færgeve ej tsvejen je Sø 61 jen 555 561 ndervan 326 326 Amtsve Ellinge g Vibehus s Nørregad n vej Overby Klint Nørrevang 326 Lyng 563 666 Od 561 555 denvej 555 5 e kvej 61 r Rørvig Sølagerveje Klintve væ 5 s 6 562 666 vej 3 66 t 6 Skansehage Klin 6 j Lumsås 3 6 6 evej Sølager 56 Nakke Lyng e Frederik Stor rlsmind ken To ldbodgad e Ka 26 561 555 Møllebak 3 326 Od 2 666 denve 56 Lynæs 5 j Kirke 61 Tengslemark Rørvigvej 326 å sve 563 666 562 666 j jen Nyrup e edvej rd j 66 Oddenvej 563 sherr 2 Vangen angsv St 561 6 vej Ve Od 66 ve en rk s 6 562 s evma 563 ter 56 555tru Te nglesl 56 lyng derv pv 562 3 v Ku ej 66 ej Rørvig Nakke No Søn 6 Nakke No Søn lhusvej n veje j k Kulhuse Bara Stenstrup ve p 316 565 Barakvejen 561 555 563 Nyru Sa 316 666 xilds Nakke Kirkeå 560 561 all 555 561 666 e Nyled 5 Nykøbing Sj. -

Dybendals Grusgrav, Dybendalvej 14A Og 16, 4190 Munke Bjergby Gældende Indtil 21.11.2026
Dybendals Grusgrav, Dybendalvej 14A og 16, 4190 Munke Bjergby Gældende indtil 21.11.2026 Matr.nr. 5h, 5i og 13b Munke Bjergby By, Munke Bjergby Dybendalvej 14A og 16, 4190 Munke Bjergby Sorø Regionale Graveområde – Bromme Delområde Dato: 24.06.2021 Sagsnummer EMN-2020-32473 Oversigt Dybendalvej 14A og 16, 4190 Munke Bjergby, Matr. 5h, 5i, og 13b Munke Bjergby By, Munke Bjergby Sagsnummer EMN-2020-32473 (tillæg til tilladelse 15/00064) Ansøger, indvinder og ejer Navn: DMG/SGS CVR-nr.: 14407677 Adresse: Løngvej 14, 4180 Sorø Kontakt: Daniel Opiola Telefon: 25272311 E-mail: [email protected] Tilladelse til indvinding af råstoffer 3 Tilladelse til indvinding af råstoffer Indhold 1 AFGØRELSER FRA REGION SJÆLLAND ................................................................................................................. 6 1.1 RÅSTOFLOVEN ....................................................................................................................................................... 6 1.1.1 Begrundelse .................................................................................................................................................. 6 2 VILKÅR ............................................................................................................................................................... 6 2.1 VILKÅR EFTER RÅSTOFLOVEN .................................................................................................................................... 6 2.1.1 Tilførsel af råstoffer .....................................................................................................................................