Ecography ECOG-04374 Chen, C., Qu, Y., Zhou, X
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Biotic and Abiotic Influences on Activity Patterns Of
Biological Conservation 97 (2001) 387±398 www.elsevier.com/locate/biocon Biotic and abiotic in¯uences on activity patterns of insular pit-vipers (Gloydius shedaoensis, Viperidae) from north-eastern China Li-xin Sun a, Richard Shine b,*, Zhao Debi a, Tang Zhengren a aSnake Island Protection District, Lushun, People's Republic of China bBiological Sciences A08, University of Sydney, NSW 2006, Australia Received 3 May 2000; received in revised form 18 July 2000; accepted 19 July 2000 Abstract In order to use counts of active animals to estimate population parameters (abundance, sex ratio, age structure), we need to understand the factors that bias such counts. For many taxa, the main problems involve behavioural dierences among age/sex classes, and the eects of local conditions on activity levels. A unique opportunity to quantify such eects on snakes occurs on Shedao, a small island in the Bohai Sea o north-eastern China. The island contains an extraordinary density of endemic pit-vipers (Gloydius shedaoensis), that feed primarily on migrating passerine birds. Over an 8-year period we walked the same 540-m path on 936 mornings during bird-migration periods, counted all pit-vipers within a 3-m-wide transect, and recorded the animals' sex and age class (adult vs juvenile). Total numbers of snakes averaged 40.6 per survey (0.31 per m): thus, the data set contains 37,980 records of sightings of snakes. The total numbers and the composition (sex ratio, age structure) of snakes seen in a morning diered among segments of the path, diered between seasons (spring versus autumn), diered with time within each season, and were in¯uenced by weather conditions (temperature, wind speed, relative humidity). -

New Records of Snakes (Squamata: Serpentes) from Hoa Binh Province, Northwestern Vietnam
Bonn zoological Bulletin 67 (1): 15–24 May 2018 New records of snakes (Squamata: Serpentes) from Hoa Binh Province, northwestern Vietnam Truong Quang Nguyen1,2,*, Tan Van Nguyen 1,3, Cuong The Pham1,2, An Vinh Ong4 & Thomas Ziegler5 1 Institute of Ecology and Biological Resources, Vietnam Academy of Science and Technology, 18 Hoang Quoc Viet Road, Hanoi, Vietnam 2 Graduate University of Science and Technology, Vietnam Academy of Science and Technology, 18 Hoang Quoc Viet Road, Hanoi, Vietnam 3 Save Vietnam’s Wildlife, Cuc Phuong National Park, Ninh Binh Province, Vietnam 4 Vinh University, 182 Le Duan Road, Vinh City, Nghe An Province, Vietnam 5 AG Zoologischer Garten Köln, Riehler Strasse 173, D-50735 Cologne, Germany * Corresponding author. E-mail: [email protected] Abstract. We report nine new records of snakes from Hoa Binh Province based on a reptile collection from Thuong Tien, Hang Kia-Pa Co, Ngoc Son-Ngo Luong nature reserves, and Tan Lac District, comprising six species of Colubri- dae (Dryocalamus davisonii, Euprepiophis mandarinus, Lycodon futsingensis, L. meridionalis, Sibynophis collaris and Sinonatrix aequifasciata), one species of Pareatidae (Pareas hamptoni) and two species of Viperidae (Protobothrops mu- crosquamatus and Trimeresurus gumprechti). In addition, we provide an updated list of 43 snake species from Hoa Binh Province. The snake fauna of Hoa Binh contains some species of conservation concern with seven species listed in the Governmental Decree No. 32/2006/ND-CP (2006), nine species listed in the Vietnam Red Data Book (2007), and three species listed in the IUCN Red List (2018). Key words. New records, snakes, taxonomy, Hoa Binh Province. -

WHO Guidance on Management of Snakebites
GUIDELINES FOR THE MANAGEMENT OF SNAKEBITES 2nd Edition GUIDELINES FOR THE MANAGEMENT OF SNAKEBITES 2nd Edition 1. 2. 3. 4. ISBN 978-92-9022- © World Health Organization 2016 2nd Edition All rights reserved. Requests for publications, or for permission to reproduce or translate WHO publications, whether for sale or for noncommercial distribution, can be obtained from Publishing and Sales, World Health Organization, Regional Office for South-East Asia, Indraprastha Estate, Mahatma Gandhi Marg, New Delhi-110 002, India (fax: +91-11-23370197; e-mail: publications@ searo.who.int). The designations employed and the presentation of the material in this publication do not imply the expression of any opinion whatsoever on the part of the World Health Organization concerning the legal status of any country, territory, city or area or of its authorities, or concerning the delimitation of its frontiers or boundaries. Dotted lines on maps represent approximate border lines for which there may not yet be full agreement. The mention of specific companies or of certain manufacturers’ products does not imply that they are endorsed or recommended by the World Health Organization in preference to others of a similar nature that are not mentioned. Errors and omissions excepted, the names of proprietary products are distinguished by initial capital letters. All reasonable precautions have been taken by the World Health Organization to verify the information contained in this publication. However, the published material is being distributed without warranty of any kind, either expressed or implied. The responsibility for the interpretation and use of the material lies with the reader. In no event shall the World Health Organization be liable for damages arising from its use. -

Species Identification of Shed Snake Skins in Taiwan and Adjacent Islands
Zoological Studies 56: 38 (2017) doi:10.6620/ZS.2017.56-38 Open Access Species Identification of Shed Snake Skins in Taiwan and Adjacent Islands Tein-Shun Tsai1,* and Jean-Jay Mao2 1Department of Biological Science and Technology, National Pingtung University of Science and Technology 1 Shuefu Road, Neipu, Pingtung 912, Taiwan 2Department of Forestry and Natural Resources, National Ilan University No.1, Sec. 1, Shennong Rd., Yilan City, Yilan County 260, Taiwan. E-mail: [email protected] (Received 28 August 2017; Accepted 25 November 2017; Published 19 December 2017; Communicated by Jian-Nan Liu) Tein-Shun Tsai and Jean-Jay Mao (2017) Shed snake skins have many applications for humans and other animals, and can provide much useful information to a field survey. When properly prepared and identified, a shed snake skin can be used as an important voucher; the morphological descriptions of the shed skins may be critical for taxonomic research, as well as studies of snake ecology and conservation. However, few convenient/ expeditious methods or techniques to identify shed snake skins in specific areas have been developed. In this study, we collected and examined a total of 1,260 shed skin samples - including 322 samples from neonates/ juveniles and 938 from subadults/adults - from 53 snake species in Taiwan and adjacent islands, and developed the first guide to identify them. To the naked eye or from scanned images, the sheds of almost all species could be identified if most of the shed was collected. The key features that aided in identification included the patterns on the sheds and scale morphology. -

NHBSS 061 1G Hikida Fieldg
Book Review N$7+IST. BULL. S,$0 SOC. 61(1): 41–51, 2015 A Field Guide to the Reptiles of Thailand by Tanya Chan-ard, John W. K. Parr and Jarujin Nabhitabhata. Oxford University Press, New York, 2015. 344 pp. paper. ISBN: 9780199736492. 7KDLUHSWLOHVZHUHÀUVWH[WHQVLYHO\VWXGLHGE\WZRJUHDWKHUSHWRORJLVWV0DOFROP$UWKXU 6PLWKDQG(GZDUG+DUULVRQ7D\ORU7KHLUFRQWULEXWLRQVZHUHSXEOLVKHGDV6MITH (1931, 1935, 1943) and TAYLOR 5HFHQWO\RWKHUERRNVDERXWUHSWLOHVDQGDPSKLELDQV LQ7KDLODQGZHUHSXEOLVKHG HJ&HAN-ARD ET AL., 1999: COX ET AL DVZHOODVPDQ\ SDSHUV+RZHYHUWKHVHERRNVZHUHWD[RQRPLFVWXGLHVDQGQRWJXLGHVIRURUGLQDU\SHRSOH7ZR DGGLWLRQDOÀHOGJXLGHERRNVRQUHSWLOHVRUDPSKLELDQVDQGUHSWLOHVKDYHDOVREHHQSXEOLVKHG 0ANTHEY & GROSSMANN, 1997; DAS EXWWKHVHERRNVFRYHURQO\DSDUWRIWKHIDXQD The book under review is very well prepared and will help us know Thai reptiles better. 2QHRIWKHDXWKRUV-DUXMLQ1DEKLWDEKDWDZDVP\ROGIULHQGIRUPHUO\WKH'LUHFWRURI1DWXUDO +LVWRU\0XVHXPWKH1DWLRQDO6FLHQFH0XVHXP7KDLODQG+HZDVDQH[FHOOHQWQDWXUDOLVW DQGKDGH[WHQVLYHNQRZOHGJHDERXW7KDLDQLPDOVHVSHFLDOO\DPSKLELDQVDQGUHSWLOHV,Q ZHYLVLWHG.KDR6RL'DR:LOGOLIH6DQFWXDU\WRVXUYH\KHUSHWRIDXQD+HDGYLVHGXV WRGLJTXLFNO\DURXQGWKHUH:HFROOHFWHGIRXUVSHFLPHQVRIDibamusZKLFKZHGHVFULEHG DVDQHZVSHFLHVDibamus somsaki +ONDA ET AL 1RZ,DPYHU\JODGWRNQRZWKDW WKLVERRNZDVSXEOLVKHGE\KLPDQGKLVFROOHDJXHV8QIRUWXQDWHO\KHSDVVHGDZD\LQ +LVXQWLPHO\GHDWKPD\KDYHGHOD\HGWKHSXEOLFDWLRQRIWKLVERRN7KHERRNLQFOXGHVQHDUO\ DOOQDWLYHUHSWLOHV PRUHWKDQVSHFLHV LQ7KDLODQGDQGPRVWSLFWXUHVZHUHGUDZQZLWK H[FHOOHQWGHWDLO,WLVDYHU\JRRGÀHOGJXLGHIRULGHQWLÀFDWLRQRI7KDLUHSWLOHVIRUVWXGHQWV -

Vol. 25 No. 1 March, 2000 H a M a D R Y a D V O L 25
NO.1 25 M M A A H D A H O V D A Y C R R L 0 0 0 2 VOL. 25NO.1 MARCH, 2000 2% 3% 2% 3% 2% 3% 2% 3% 2% 3% 2% 3% 2% 3% 2% 3% 2% 3% 4% 5% 4% 5% 4% 5% 4% 5% 4% 5% 4% 5% 4% 5% 4% 5% 4% 5% HAMADRYAD Vol. 25. No. 1. March 2000 Date of issue: 31 March 2000 ISSN 0972-205X Contents A. E. GREER & D. G. BROADLEY. Six characters of systematic importance in the scincid lizard genus Mabuya .............................. 1–12 U. MANTHEY & W. DENZER. Description of a new genus, Hypsicalotes gen. nov. (Sauria: Agamidae) from Mt. Kinabalu, North Borneo, with remarks on the generic identity of Gonocephalus schultzewestrumi Urban, 1999 ................13–20 K. VASUDEVAN & S. K. DUTTA. A new species of Rhacophorus (Anura: Rhacophoridae) from the Western Ghats, India .................21–28 O. S. G. PAUWELS, V. WALLACH, O.-A. LAOHAWAT, C. CHIMSUNCHART, P. DAVID & M. J. COX. Ethnozoology of the “ngoo-how-pak-pet” (Serpentes: Typhlopidae) in southern peninsular Thailand ................29–37 S. K. DUTTA & P. RAY. Microhyla sholigari, a new species of microhylid frog (Anura: Microhylidae) from Karnataka, India ....................38–44 Notes R. VYAS. Notes on distribution and breeding ecology of Geckoella collegalensis (Beddome, 1870) ..................................... 45–46 A. M. BAUER. On the identity of Lacerta tjitja Ljungh 1804, a gecko from Java .....46–49 M. F. AHMED & S. K. DUTTA. First record of Polypedates taeniatus (Boulenger, 1906) from Assam, north-eastern India ...................49–50 N. M. ISHWAR. Melanobatrachus indicus Beddome, 1878, resighted at the Anaimalai Hills, southern India ............................. -

Lotus Wind Power Project
Initial Environmental Examination – Appendix H Project Number: 54211-001 March 2021 Document Stage: Draft Viet Nam: Lotus Wind Power Project Prepared by ERM Vietnam for Lien Lap Wind Power Joint Stock Company, Phong Huy Wind Power Joint Stock Company, and Phong Nguyen Wind Power Joint Stock Company as a requirement of the Asian Development Bank. The initial environmental examination is a document of the borrower. The views expressed herein do not necessarily represent those of ADB's Board of Directors, Management, or staff, and may be preliminary in nature. Your attention is directed to the “Terms of Use” section of this website. In preparing any country program or strategy, financing any project, or by making any designation of or reference to a particular territory or geographic area in this document, the Asian Development Bank does not intend to make any judgments as to the legal or other status of any territory or area. Biodiversity survey Wet season report Phong Huy Wind Power Project, Huong Hoa, Quang Tri, Viet Nam 7 July 2020 Prepared by ERM’s Subcontractor for ERM Vietnam Document details Document title Biodiversity survey Wet season report Document subtitle Phong Huy Wind Power Project, Huong Hoa, Quang Tri, Viet Nam Date 7 July 2020 Version 1.0 Author ERM’s Subcontractor Client Name ERM Vietnam Document history Version Revision Author Reviewed by ERM approval to issue Comments Name Date Draft 1.0 Name Name Name 00.00.0000 Text Version: 1.0 Client: ERM Vietnam 7 July 2020 BIODIVERSITY SURVEY WET SEASON REPORT CONTENTS Phong Huy Wind Power Project, Huong Hoa, Quang Tri, Viet Nam CONTENTS 1. -

Russell's Viper (Daboia Russelii) in Bangladesh: Its Boom and Threat To
J. Asiat. Soc. Bangladesh, Sci. 44(1): 15-22, June 2018 RUSSELL’S VIPER (DABOIA RUSSELII) IN BANGLADESH: ITS BOOM AND THREAT TO HUMAN LIFE MD. FARID AHSAN1* AND MD. ABU SAEED2 1Department of Zoology, University of Chittagong, Chittagong, Bangladesh 2 555, Kazipara, Mirpur, Dhaka-1216, Bangladesh Abstract The occurrence of Russell’s viper (Daboia russelii Shaw and Nodder 1797) in Bangladesh is century old information and its rarity was known to the wildlife biologists till 2013 but its recent booming is also causing a major threat to human life in the area. Recently it has been reported from nine districts (Dinajpur, Chapai Nawabganj, Rajshahi, Naogaon, Natore, Pabna, Rajbari, Chuadanga and Patuakhali) and old records revealed 11 districts (Nilphamari, Dinajpur, Rangpur, Chapai Nawabganj, Rajshahi, Bogra, Jessore, Satkhira, Khulna, Bagerhat and Chittagong). Thus altogether 17 out of 64 districts in Bangladesh, of which Chapai Nawabganj and Rajshahi are most affected and 20 people died due to Russell’s viper bite during 2013 to 2016. Its past and present distribution in Bangladesh and death toll of its bites have been discussed. Its booming causes have also been predicted and precautions have been recommended. Research on Russell’s viper is deemed necessary due to reemergence in deadly manner. Key words: Russell’s viper, Daboia russelii, Distribution, Boom, Panic, Death toll Introduction Two species of Russell’s viper are known to occur in this universe of which Daboia russelii (Shaw and Nodder 1797) is distributed in Pakistan, India, Nepal, Bhutan, Bangladesh and Sri Lanka (www.reptile.data-base.org); while Daboia siamensis (Smith 1917) occurs in China, Myanmar, Indonesia, Thailand, Taiwan and Cambodia (Wogan 2012). -

Proceedings of the United States National Museum
CHINESE AMPHIBIANS AND KEPTILES IN THE UNITED STATES NATIONAL MUSEUM By Leonhard Stejneger Head Curator of Biology, United States National Museum The United States National Museum has of late years received a lar^e number of amphibians and reptiles from China, and as there are indications of an increasing interest in these vertebrates among students and collectors in that country, it has been thought best to give as full an account as practicable of the material available, in order to acquaint them not only with what the National Museum possesses, but inferentially with what is still needed before a com- plete herpetology of Qhina proper can be attempted. In my Herpetology of Japan and Adjacent Territory^ I included not only the Russian Coast Province of Siberia, but also Korea, eastern Manchuria, and adjoining parts of northeastern China proper. Each genus and species, the Chinese ones included, were treated in detail, with full synonymy and bibliographic references, etc. In a subsequent paper, The Batrachians and Keptiles of Formosa,- genera and species added to the fauna since 1907 were similarly elaborated in full. It has been considered unnecessary to repeat these synony- mies and references in the present paper, hence only genera and species not found in the two earlier works are here treated in the same manner. However, reference has been given under each species to the page in the Herpetology of Japan and the paper re- lating to Formosa, where the species with synonymy, description, and frequently also illustration may be found. In addition, refer- ences omitted in the earlier work are given in so far as they relate to China proper, but no attempt has been made to include those re- ferring to Hainan and Formosa. -

Evaluation of the Effectiveness of Three Survey Methods for Sampling Terrestrial Herpetofauna in South China
Herpetological Conservation and Biology 6(3):479–489. Submitted: 9 July 2011, Accepted: 16 October 2011 Published: 31 December 2011. EVALUATION OF THE EFFECTIVENESS OF THREE SURVEY METHODS FOR SAMPLING TERRESTRIAL HERPETOFAUNA IN SOUTH CHINA 1 YIK-HEI SUNG, NANCY E. KARRAKER, AND BILLY C.H. HAU School of Biological Sciences, The University of Hong Kong, Pokfulam Road, Hong Kong SAR, China 1Corresponding author, email: [email protected] Abstract.—Southeast Asia exhibits high herpetofaunal biodiversity, yet many areas and taxa in the region remain understudied. Extensive surveys are needed to fill information gaps, yet at present we have little knowledge about the effectiveness of different herpetofaunal survey methods in the region. We conducted field studies to examine the effectiveness of three survey methods for sampling terrestrial amphibians and reptiles in Hong Kong. Transect surveys were the most effective at sampling species richness and drift fences with pitfall traps and funnel traps were the most efficient in capturing high numbers of reptiles. We recommend the use of transect surveys for rapid biodiversity assessment and the combination of transect surveys and pitfall traps for comprehensive species inventories. Pitfall traps represent an excellent tool for surveys or population monitoring of leaf litter species. The results of this study will aid researchers in assessing the feasibility of and in choosing herpetofaunal survey methods in Southeast Asia. Key Words.—amphibians; coverboards; drift fences; monitoring; pitfall traps; reptiles; species inventories; transect surveys INTRODUCTION applied (Gardner et al. 2008). Therefore, successful studies must employ survey methods that permit the Many amphibian and reptile populations are declining most efficient completion of study objectives (Ribeiro- at unprecedented rates and some risk extinction under Junior et al. -
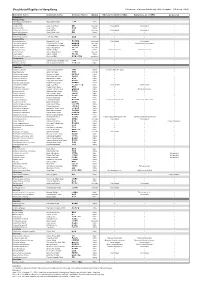
Checklist of Reptiles in Hong Kong © Programme of Ecology & Biodiversity, HKU (Last Update: 10 September 2012)
Checklist of Reptiles in Hong Kong © Programme of Ecology & Biodiversity, HKU (Last Update: 10 September 2012) Scientific name Common name Chinese Name Status Ades & Kendrick (2004) Karsen et al. (1998) Uetz et al. Testudines Platysternidae Platysternon megacephalum Big-headed Terrapin 大頭龜 Native Cheloniidae Caretta caretta Loggerhead Turtle 蠵龜 Uncertain Not included Not included Chelonia mydas Green Turtle 緣海龜 Native Eretmochelys imbricata Hawksbill Turtle 玳瑁 Uncertain Not included Not included Lepidochelys olivacea Pacific Ridley Turtle 麗龜 Native Dermochelyidae Dermochelys coriacea Leatherback Turtle 稜皮龜 Native Geoemydidae Cuora amboinensis Malayan Box Turtle 馬來閉殼龜 Introduced Not included Not included Cuora flavomarginata Yellow-lined Box Terrapin 黃緣閉殼龜 Uncertain Cistoclemmys flavomarginata Cuora trifasciata Three-banded Box Terrapin 三線閉殼龜 Native Mauremys mutica Chinese Pond Turtle 黃喉水龜 Uncertain Mauremys reevesii Reeves' Terrapin 烏龜 Native Chinemys reevesii Chinemys reevesii Ocadia sinensis Chinese Striped Turtle 中華花龜 Uncertain Sacalia bealei Beale's Terrapin 眼斑水龜 Native Trachemys scripta elegans Red-eared Slider 巴西龜 / 紅耳龜 Introduced Trionychidae Palea steindachneri Wattle-necked Soft-shelled Turtle 山瑞鱉 Uncertain Pelodiscus sinensis Chinese Soft-shelled Turtle 中華鱉 / 水魚 Native Squamata - Serpentes Colubridae Achalinus rufescens Rufous Burrowing Snake 棕脊蛇 Native Achalinus refescens (typo) Ahaetulla prasina Jade Vine Snake 綠瘦蛇 Uncertain Amphiesma atemporale Mountain Keelback 無顳鱗游蛇 Native -

P. 1 AC27 Inf. 7 (English Only / Únicamente En Inglés / Seulement
AC27 Inf. 7 (English only / únicamente en inglés / seulement en anglais) CONVENTION ON INTERNATIONAL TRADE IN ENDANGERED SPECIES OF WILD FAUNA AND FLORA ____________ Twenty-seventh meeting of the Animals Committee Veracruz (Mexico), 28 April – 3 May 2014 Species trade and conservation IUCN RED LIST ASSESSMENTS OF ASIAN SNAKE SPECIES [DECISION 16.104] 1. The attached information document has been submitted by IUCN (International Union for Conservation of * Nature) . It related to agenda item 19. * The geographical designations employed in this document do not imply the expression of any opinion whatsoever on the part of the CITES Secretariat or the United Nations Environment Programme concerning the legal status of any country, territory, or area, or concerning the delimitation of its frontiers or boundaries. The responsibility for the contents of the document rests exclusively with its author. AC27 Inf. 7 – p. 1 Global Species Programme Tel. +44 (0) 1223 277 966 219c Huntingdon Road Fax +44 (0) 1223 277 845 Cambridge CB3 ODL www.iucn.org United Kingdom IUCN Red List assessments of Asian snake species [Decision 16.104] 1. Introduction 2 2. Summary of published IUCN Red List assessments 3 a. Threats 3 b. Use and Trade 5 c. Overlap between international trade and intentional use being a threat 7 3. Further details on species for which international trade is a potential concern 8 a. Species accounts of threatened and Near Threatened species 8 i. Euprepiophis perlacea – Sichuan Rat Snake 9 ii. Orthriophis moellendorfi – Moellendorff's Trinket Snake 9 iii. Bungarus slowinskii – Red River Krait 10 iv. Laticauda semifasciata – Chinese Sea Snake 10 v.