Genes Associated with Anhedonia
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Mouse Germ Line Mutations Due to Retrotransposon Insertions Liane Gagnier1, Victoria P
Gagnier et al. Mobile DNA (2019) 10:15 https://doi.org/10.1186/s13100-019-0157-4 REVIEW Open Access Mouse germ line mutations due to retrotransposon insertions Liane Gagnier1, Victoria P. Belancio2 and Dixie L. Mager1* Abstract Transposable element (TE) insertions are responsible for a significant fraction of spontaneous germ line mutations reported in inbred mouse strains. This major contribution of TEs to the mutational landscape in mouse contrasts with the situation in human, where their relative contribution as germ line insertional mutagens is much lower. In this focussed review, we provide comprehensive lists of TE-induced mouse mutations, discuss the different TE types involved in these insertional mutations and elaborate on particularly interesting cases. We also discuss differences and similarities between the mutational role of TEs in mice and humans. Keywords: Endogenous retroviruses, Long terminal repeats, Long interspersed elements, Short interspersed elements, Germ line mutation, Inbred mice, Insertional mutagenesis, Transcriptional interference Background promoter and polyadenylation motifs and often a splice The mouse and human genomes harbor similar types of donor site [10, 11]. Sequences of full-length ERVs can TEs that have been discussed in many reviews, to which encode gag, pol and sometimes env, although groups of we refer the reader for more in depth and general infor- LTR retrotransposons with little or no retroviral hom- mation [1–9]. In general, both human and mouse con- ology also exist [6–9]. While not the subject of this re- tain ancient families of DNA transposons, none view, ERV LTRs can often act as cellular enhancers or currently active, which comprise 1–3% of these genomes promoters, creating chimeric transcripts with genes, and as well as many families or groups of retrotransposons, have been implicated in other regulatory functions [11– which have caused all the TE insertional mutations in 13]. -

Splicing Regulatory Factors in Breast Cancer Hallmarks and Disease Progression
www.oncotarget.com Oncotarget, 2019, Vol. 10, (No. 57), pp: 6021-6037 Review Splicing regulatory factors in breast cancer hallmarks and disease progression Esmee Koedoot1, Liesanne Wolters1, Bob van de Water1 and Sylvia E. Le Dévédec1 1Division of Drug Discovery and Safety, LACDR, Leiden University, Leiden, The Netherlands Correspondence to: Sylvia E. Le Dévédec, email: [email protected] Keywords: hallmarks of cancer; breast cancer; alternative splicing; splice factors; RNA sequencing Received: April 23, 2019 Accepted: August 29, 2019 Published: October 15, 2019 Copyright: Koedoot et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License 3.0 (CC BY 3.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. ABSTRACT By regulating transcript isoform expression levels, alternative splicing provides an additional layer of protein control. Recent studies show evidence that cancer cells use different splicing events to fulfill their requirements in order to develop, progress and metastasize. However, there has been less attention for the role of the complex catalyzing the complicated multistep splicing reaction: the spliceosome. The spliceosome consists of multiple sub-complexes in total comprising 244 proteins or splice factors and 5 associated RNA molecules. Here we discuss the role of splice factors in the oncogenic processes tumors cells need to fulfill their oncogenic properties (the so-called the hallmarks of cancer). Despite the fact that splice factors have been investigated only recently, they seem to play a prominent role in already five hallmarks of cancer: angiogenesis, resisting cell death, sustaining proliferation, deregulating cellular energetics and invasion and metastasis formation by affecting major signaling pathways such as epithelial-to-mesenchymal transition, the Warburg effect, DNA damage response and hormone receptor dependent proliferation. -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

Supplementary Table 1: Adhesion Genes Data Set
Supplementary Table 1: Adhesion genes data set PROBE Entrez Gene ID Celera Gene ID Gene_Symbol Gene_Name 160832 1 hCG201364.3 A1BG alpha-1-B glycoprotein 223658 1 hCG201364.3 A1BG alpha-1-B glycoprotein 212988 102 hCG40040.3 ADAM10 ADAM metallopeptidase domain 10 133411 4185 hCG28232.2 ADAM11 ADAM metallopeptidase domain 11 110695 8038 hCG40937.4 ADAM12 ADAM metallopeptidase domain 12 (meltrin alpha) 195222 8038 hCG40937.4 ADAM12 ADAM metallopeptidase domain 12 (meltrin alpha) 165344 8751 hCG20021.3 ADAM15 ADAM metallopeptidase domain 15 (metargidin) 189065 6868 null ADAM17 ADAM metallopeptidase domain 17 (tumor necrosis factor, alpha, converting enzyme) 108119 8728 hCG15398.4 ADAM19 ADAM metallopeptidase domain 19 (meltrin beta) 117763 8748 hCG20675.3 ADAM20 ADAM metallopeptidase domain 20 126448 8747 hCG1785634.2 ADAM21 ADAM metallopeptidase domain 21 208981 8747 hCG1785634.2|hCG2042897 ADAM21 ADAM metallopeptidase domain 21 180903 53616 hCG17212.4 ADAM22 ADAM metallopeptidase domain 22 177272 8745 hCG1811623.1 ADAM23 ADAM metallopeptidase domain 23 102384 10863 hCG1818505.1 ADAM28 ADAM metallopeptidase domain 28 119968 11086 hCG1786734.2 ADAM29 ADAM metallopeptidase domain 29 205542 11085 hCG1997196.1 ADAM30 ADAM metallopeptidase domain 30 148417 80332 hCG39255.4 ADAM33 ADAM metallopeptidase domain 33 140492 8756 hCG1789002.2 ADAM7 ADAM metallopeptidase domain 7 122603 101 hCG1816947.1 ADAM8 ADAM metallopeptidase domain 8 183965 8754 hCG1996391 ADAM9 ADAM metallopeptidase domain 9 (meltrin gamma) 129974 27299 hCG15447.3 ADAMDEC1 ADAM-like, -

Aneuploidy: Using Genetic Instability to Preserve a Haploid Genome?
Health Science Campus FINAL APPROVAL OF DISSERTATION Doctor of Philosophy in Biomedical Science (Cancer Biology) Aneuploidy: Using genetic instability to preserve a haploid genome? Submitted by: Ramona Ramdath In partial fulfillment of the requirements for the degree of Doctor of Philosophy in Biomedical Science Examination Committee Signature/Date Major Advisor: David Allison, M.D., Ph.D. Academic James Trempe, Ph.D. Advisory Committee: David Giovanucci, Ph.D. Randall Ruch, Ph.D. Ronald Mellgren, Ph.D. Senior Associate Dean College of Graduate Studies Michael S. Bisesi, Ph.D. Date of Defense: April 10, 2009 Aneuploidy: Using genetic instability to preserve a haploid genome? Ramona Ramdath University of Toledo, Health Science Campus 2009 Dedication I dedicate this dissertation to my grandfather who died of lung cancer two years ago, but who always instilled in us the value and importance of education. And to my mom and sister, both of whom have been pillars of support and stimulating conversations. To my sister, Rehanna, especially- I hope this inspires you to achieve all that you want to in life, academically and otherwise. ii Acknowledgements As we go through these academic journeys, there are so many along the way that make an impact not only on our work, but on our lives as well, and I would like to say a heartfelt thank you to all of those people: My Committee members- Dr. James Trempe, Dr. David Giovanucchi, Dr. Ronald Mellgren and Dr. Randall Ruch for their guidance, suggestions, support and confidence in me. My major advisor- Dr. David Allison, for his constructive criticism and positive reinforcement. -

Epigenetic Regulation of Murine Stab2 Expression in Determining Atherosclerotic Susceptibility
EPIGENETIC REGULATION OF MURINE STAB2 EXPRESSION IN DETERMINING ATHEROSCLEROTIC SUSCEPTIBILITY Sharlene Dong A thesis submitted to the faculty of the University of North Carolina at Chapel Hill in partial fulfillment of the requirements for the degree of Master of Science in the Department of Nutrition (Nutritional Biochemistry) in the Gillings School of Global Public Health Chapel Hill 2017 Approved by: Nobuyo Maeda Rosalind Coleman Eric L. Klett i © 2017 Sharlene Dong ALL RIGHTS RESERVED ii ABSTRACT Sharlene Dong: Epigenetic regulation of murine Stab2 expression in determining atherosclerotic susceptibility (Under the direction of Nobuyo Maeda) Atherosclerosis is a progressive disease characterized by the accumulation of lipids and fibrous elements in the arteries. In mice, the aortic arch and root show distinct patterns of lesion development. A quantitative trait locus (QTL) analysis of an intercross between apoE-null mice on 129S6 and DBA/2J backgrounds has revealed a significant locus on chromosome 10, Aath5, that affects plaque size in the arch. One candidate gene in the Aath5 locus is the Stab2 gene, which encodes a receptor for hyaluronan. The DBA allele of the Stab2 gene includes a unique intracisternal A-particle (IAP) retrotransposon inserted in reverse in the promoter region, which drives Stab2 expression. Methylation status of this region was analyzed through allele-specific bisulfite sequencing and correlated with ectopic Stab2 expression in the heart and kidneys. Thus, epigenetic regulation of this IAP element may play a role in regulating Stab2 expression and affect atherosclerotic susceptibility in the aortic arch. iii To my mom and dad and all my friends Sincerely thank you for your unwavering support in me. -
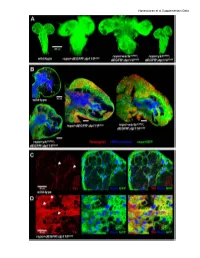
Supplementary Data Vigneswaran Et Al Supplementary Data
Vigneswaran et al Supplementary Data Vigneswaran et al Supplementary Data Figure S1: Yki is required for EGFR-PI3K-driven glial neoplasia in Drosophila (A) Optical projections of whole brain-nerve cord complexes from 3rd instar larvae approximately 130 hrs old. Dorsal view; anterior up. CD8-GFP (green) labels glial cell bodies. Compared to repo>dEGFRλ;dp110CAAX, warts knockdown (repo>wartsdsRNA; dEGFRλ;dp110CAAX) increased neoplastic brain overgrowth and yki knockdown (repo>ykidsRNA;dEGFRλ;dp110CAAX) decreased neoplastic brain overgrowth. (B) 3 µm optical projections of brain hemispheres, age-matched 3rd instar larvae. Frontal sections; anterior up; midline to left. Repo (red) labels glial cell nuclei; CD8-GFP (green) labels glial cell bodies; anti-HRP (blue) counter-stains for neurons and neuropil. (middle) repo>dEGFRλ;dp110CAAX showed increased glial cell numbers (red nuclei) compared to (upper left) wild-type. Compared to repo>dEGFRλ;dp110CAAX, (right) warts knockdown increased neoplastic glial cell numbers (red nuclei), whereas (lower left) yki knockdown reduced neoplastic glial cell numbers (red nuclei). (C, D) Low levels of Yki protein (red) was observed in wild-type central brain glia (white arrows, left panel in C) compared to high levels of cytoplasmic and nuclear Yki protein in dEGFRλ;dp110CAAX neoplastic glia (white arrows, left panel in D); Repo (blue) labels glial cell nuclei; CD8-GFP (green) labels glial cell bodies. Vigneswaran et al Supplementary Data Figure S2: YAP/TAZ expression confined to RTK-amplified tuMor cells and Maintained in patient-derived xenografts (A) On the left, immunohistochemical (IHC) staining in representative normal brain parenchyma in the cortex where YAP expression and TAZ expression was limited to vascular cells and was not detectable in normal neuronal and glial cells. -

Crystal Structure of a Variant PAM2 Motif of LARP4B Bound to the MLLE Domain of PABPC1
biomolecules Article Crystal Structure of a Variant PAM2 Motif of LARP4B Bound to the MLLE Domain of PABPC1 Clemens Grimm *, Jann-Patrick Pelz, Cornelius Schneider, Katrin Schäffler and Utz Fischer Department of Biochemistry, Theodor Boveri Institute, Biocenter of the University of Würzburg, 97070 Würzburg, Germany; [email protected] (J.-P.P.); [email protected] (C.S.); katrin.schaeffl[email protected] (K.S.); utz.fi[email protected] (U.F.) * Correspondence: [email protected] Received: 3 April 2020; Accepted: 4 June 2020; Published: 6 June 2020 Abstract: Eukaryotic cells determine the protein output of their genetic program by regulating mRNA transcription, localization, translation and turnover rates. This regulation is accomplished by an ensemble of RNA-binding proteins (RBPs) that bind to any given mRNA, thus forming mRNPs. Poly(A) binding proteins (PABPs) are prominent members of virtually all mRNPs that possess poly(A) tails. They serve as multifunctional scaffolds, allowing the recruitment of diverse factors containing a poly(A)-interacting motif (PAM) into mRNPs. We present the crystal structure of the variant PAM motif (termed PAM2w) in the N-terminal part of the positive translation factor LARP4B, which binds to the MLLE domain of the poly(A) binding protein C1 cytoplasmic 1 (PABPC1). The structural analysis, along with mutational studies in vitro and in vivo, uncovered a new mode of interaction between PAM2 motifs and MLLE domains. Keywords: PAM2w; PAM2; PABC1; MLLE domain; PABP; Poly(A) binding protein 1. Introduction In higher eukaryotes, posttranscriptional mechanisms contribute significantly to gene expression. The mRNA, which is translated into proteins by the ribosomes, has first to undergo several maturation steps, from the primary pre-mRNA transcript to the mature mRNA. -

Open Data for Differential Network Analysis in Glioma
International Journal of Molecular Sciences Article Open Data for Differential Network Analysis in Glioma , Claire Jean-Quartier * y , Fleur Jeanquartier y and Andreas Holzinger Holzinger Group HCI-KDD, Institute for Medical Informatics, Statistics and Documentation, Medical University Graz, Auenbruggerplatz 2/V, 8036 Graz, Austria; [email protected] (F.J.); [email protected] (A.H.) * Correspondence: [email protected] These authors contributed equally to this work. y Received: 27 October 2019; Accepted: 3 January 2020; Published: 15 January 2020 Abstract: The complexity of cancer diseases demands bioinformatic techniques and translational research based on big data and personalized medicine. Open data enables researchers to accelerate cancer studies, save resources and foster collaboration. Several tools and programming approaches are available for analyzing data, including annotation, clustering, comparison and extrapolation, merging, enrichment, functional association and statistics. We exploit openly available data via cancer gene expression analysis, we apply refinement as well as enrichment analysis via gene ontology and conclude with graph-based visualization of involved protein interaction networks as a basis for signaling. The different databases allowed for the construction of huge networks or specified ones consisting of high-confidence interactions only. Several genes associated to glioma were isolated via a network analysis from top hub nodes as well as from an outlier analysis. The latter approach highlights a mitogen-activated protein kinase next to a member of histondeacetylases and a protein phosphatase as genes uncommonly associated with glioma. Cluster analysis from top hub nodes lists several identified glioma-associated gene products to function within protein complexes, including epidermal growth factors as well as cell cycle proteins or RAS proto-oncogenes. -

Interstitial Deletion of 6Q25.2&Ndash
European Journal of Human Genetics (2009) 17, 573 – 581 & 2009 Macmillan Publishers Limited All rights reserved 1018-4813/09 $32.00 www.nature.com/ejhg ARTICLE Interstitial deletion of 6q25.2–q25.3: a novel microdeletion syndrome associated with microcephaly, developmental delay, dysmorphic features and hearing loss Sandesh Chakravarthy Sreenath Nagamani1, Ayelet Erez1, Christine Eng1, Zhishuo Ou1, Craig Chinault1, Laura Workman2, James Coldwell3, Pawel Stankiewicz1, Ankita Patel1, James R Lupski1,4,5 and Sau Wai Cheung*,1 1Department of Molecular and Human Genetics, Baylor College of Medicine, Houston, TX, USA; 2Sutter Medical Center, Sacramento, CA, USA; 3Children’s Medical Center, Tulsa, OK, USA; 4Department of Pediatrics, Baylor College of Medicine, Houston, TX, USA; 5Texas Children’s Hospital, Houston, TX, USA Interstitial deletions of 6q are rare. We report a detailed clinical and molecular characterization of four patients with interstitial deletion involving 6q25. All of our patients presented with microcephaly, developmental delay, dysmorphic features and hearing loss, whereas two of them had agenesis of the corpus callosum. We determined the size, extent and genomic content of the deletions using high-density array-comparative genomic hybridization (a-CGH), and found that a common segment spanning 3.52 Mb within the 6q25.2–q25.3 region was deleted in all four cases. We hypothesize that a subset of genes in the commonly deleted region are dosage sensitive and that haploinsufficieny of these genes impairs normal development of the brain and hearing. European Journal of Human Genetics (2009) 17, 573–581; doi:10.1038/ejhg.2008.220; published online 26 November 2008 Keywords: 6q deletion; hearing loss; microcephaly, developmental delay; agenesis of the corpus callosum; array-CGH Introduction lips, a lower frequency of microcephaly, micrognathia Since the first reported case in 1975,1 over 65 cases of and heart malformations. -

Human Induced Pluripotent Stem Cell–Derived Podocytes Mature Into Vascularized Glomeruli Upon Experimental Transplantation
BASIC RESEARCH www.jasn.org Human Induced Pluripotent Stem Cell–Derived Podocytes Mature into Vascularized Glomeruli upon Experimental Transplantation † Sazia Sharmin,* Atsuhiro Taguchi,* Yusuke Kaku,* Yasuhiro Yoshimura,* Tomoko Ohmori,* ‡ † ‡ Tetsushi Sakuma, Masashi Mukoyama, Takashi Yamamoto, Hidetake Kurihara,§ and | Ryuichi Nishinakamura* *Department of Kidney Development, Institute of Molecular Embryology and Genetics, and †Department of Nephrology, Faculty of Life Sciences, Kumamoto University, Kumamoto, Japan; ‡Department of Mathematical and Life Sciences, Graduate School of Science, Hiroshima University, Hiroshima, Japan; §Division of Anatomy, Juntendo University School of Medicine, Tokyo, Japan; and |Japan Science and Technology Agency, CREST, Kumamoto, Japan ABSTRACT Glomerular podocytes express proteins, such as nephrin, that constitute the slit diaphragm, thereby contributing to the filtration process in the kidney. Glomerular development has been analyzed mainly in mice, whereas analysis of human kidney development has been minimal because of limited access to embryonic kidneys. We previously reported the induction of three-dimensional primordial glomeruli from human induced pluripotent stem (iPS) cells. Here, using transcription activator–like effector nuclease-mediated homologous recombination, we generated human iPS cell lines that express green fluorescent protein (GFP) in the NPHS1 locus, which encodes nephrin, and we show that GFP expression facilitated accurate visualization of nephrin-positive podocyte formation in -

Loci for Phenotypic Dispersion in Plasma Leptin in Mouse Models of Diabetes Mellitus
RESEARCH ARTICLE `Fat's chances': Loci for phenotypic dispersion in plasma leptin in mouse models of diabetes mellitus Guy M. L. PerryID* Department of Biology, University of Prince Edward Island, Charlottetown, PEI, Canada * [email protected] a1111111111 a1111111111 a1111111111 Abstract a1111111111 Background a1111111111 Leptin, a critical mediator of feeding, metabolism and diabetes, is expressed on an incidental basis according to satiety. The genetic regulation of leptin should similarly be episodic. Methodology OPEN ACCESS Data from three mouse cohorts hosted by the Jackson Laboratory± 402 (174F, 228M) F Citation: Perry GML (2019) `Fat's chances': Loci 2 for phenotypic dispersion in plasma leptin in Dilute Brown non-Agouti (DBA/2)×DU6i intercrosses, 142 Non Obese Diabetic (NOD/ g7 mouse models of diabetes mellitus. PLoS ONE 14 ShiLtJ×(NOD/ShiLtJ×129S1/SvImJ.H2 ) N2 backcross females, and 204 male Nonobese (10): e0222654. https://doi.org/10.1371/journal. Nondiabetic (NON)×New Zealand Obese (NZO/HlLtJ) reciprocal backcrosses±were used pone.0222654 to test for loci associated with absolute residuals in plasma leptin and arcsin-transformed Editor: Clemens FuÈrnsinn, Medical University of percent fat (`phenotypic dispersion'; PDpLep and PDAFP). Individual data from 1,780 mice Vienna, AUSTRIA from 43 inbred strains was also used to estimate genetic variances and covariances for dis- Received: April 1, 2019 persion in each trait. Accepted: September 4, 2019 Published: October 29, 2019 Principal findings Copyright: © 2019 Guy M. L. Perry. This is an open Several loci for PDpLep were detected, including possibly syntenic Chr 17 loci, but there was access article distributed under the terms of the only a single position on Chr 6 for PDAFP.