Modeling Neuron–Glia Interactions with the Brian 2 Simulator
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Oligodendrocytes in Development, Myelin Generation and Beyond
cells Review Oligodendrocytes in Development, Myelin Generation and Beyond Sarah Kuhn y, Laura Gritti y, Daniel Crooks and Yvonne Dombrowski * Wellcome-Wolfson Institute for Experimental Medicine, Queen’s University Belfast, Belfast BT9 7BL, UK; [email protected] (S.K.); [email protected] (L.G.); [email protected] (D.C.) * Correspondence: [email protected]; Tel.: +0044-28-9097-6127 These authors contributed equally. y Received: 15 October 2019; Accepted: 7 November 2019; Published: 12 November 2019 Abstract: Oligodendrocytes are the myelinating cells of the central nervous system (CNS) that are generated from oligodendrocyte progenitor cells (OPC). OPC are distributed throughout the CNS and represent a pool of migratory and proliferative adult progenitor cells that can differentiate into oligodendrocytes. The central function of oligodendrocytes is to generate myelin, which is an extended membrane from the cell that wraps tightly around axons. Due to this energy consuming process and the associated high metabolic turnover oligodendrocytes are vulnerable to cytotoxic and excitotoxic factors. Oligodendrocyte pathology is therefore evident in a range of disorders including multiple sclerosis, schizophrenia and Alzheimer’s disease. Deceased oligodendrocytes can be replenished from the adult OPC pool and lost myelin can be regenerated during remyelination, which can prevent axonal degeneration and can restore function. Cell population studies have recently identified novel immunomodulatory functions of oligodendrocytes, the implications of which, e.g., for diseases with primary oligodendrocyte pathology, are not yet clear. Here, we review the journey of oligodendrocytes from the embryonic stage to their role in homeostasis and their fate in disease. We will also discuss the most common models used to study oligodendrocytes and describe newly discovered functions of oligodendrocytes. -

Build a Neuron
Build a Neuron Objectives: 1. To understand what a neuron is and what it does 2. To understand the anatomy of a neuron in relation to function This activity is great for ALL ages-even college students!! Materials: pipe cleaners (2 full size, 1 cut into 3 for each student) pony beads (6/student Introduction: Little kids: ask them where their brain is (I point to my head and torso areas till they shake their head yes) Talk about legos being the building blocks for a tower and relate that to neurons being the building blocks for your brain and that neurons send messages to other parts of your brain and to and from all your body parts. Give examples: touch from body to brain, movement from brain to body. Neurons are the building blocks of the brain that send and receive messages. Neurons come in all different shapes. Experiment: 1. First build soma by twisting a pipe cleaner into a circle 2. Then put a 2nd pipe cleaner through the circle and bend it over and twist the two strands together to make it look like a lollipop (axon) 3. take 3 shorter pipe cleaners attach to cell body to make dendrites 4. add 6 beads on the axon making sure there is space between beads for the electricity to “jump” between them to send the signal super fast. (myelin sheath) 5. Twist the end of the axon to make it look like 2 feet for the axon terminal. 6. Make a brain by having all of the neurons “talk” to each other (have each student hold their neuron because they’ll just throw them on a table for you to do it.) messages come in through the dendrites and if its a strong enough electrical change, then the cell body sends the Build a Neuron message down it’s axon where a neurotransmitter is released. -

Neurons – Is a Basic Cell of the Nervous System. • Neurons Carry Nerve Messages, Or Impulses, from One Part of the Body to Another
Nervous System Nerves and Nerve Cells: Neurons – is a basic cell of the nervous system. • Neurons carry nerve messages, or impulses, from one part of the body to another. Structure of a Nerve Cell: A neuron has three basic parts: 1. Body – controls the cell’s growth 2. Axon – is a long thin fiber that carries impulses away from the cell body Myelin – is a fatty material that insulates the axon and increases the speed at which an impulse travels 3. Dendrites – are short, branching fibers that carry nerve impulses toward the cell body. • A nerve impulse begins when the dendrites are stimulated. The impulse travels along the dendrites to the cell body, and then away from the cell body on the axon. • The impulse must cross a synapse to a muscle or another neuron. Synapse – is the space between an axon and the structure with which the neuron communicates. Types of Nerve Cells: Sensory Neurons – pick up information about your external and internal environment from your sense organs and your body Motor Neurons – sends impulses to your muscles and glands, causing them to react Interneurons – are located only in the brain and spinal cord, pass impulses from one neuron to another The Central Nervous System: • The nervous system consists of two parts. Your brain and spinal cord make up one part, which is called the central nervous system. 1 • The peripheral nervous system, which is the other part, is made up of all the nerves that connect the brain and spinal cord to other parts of the body. The Brain: Brain ± a moist, spongy organ weighing about three pounds is made up of billions of neurons that control almost everything you do and experience. -
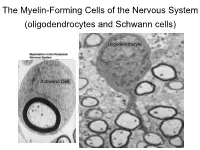
The Myelin-Forming Cells of the Nervous System (Oligodendrocytes and Schwann Cells)
The Myelin-Forming Cells of the Nervous System (oligodendrocytes and Schwann cells) Oligodendrocyte Schwann Cell Oligodendrocyte function Saltatory (jumping) nerve conduction Oligodendroglia PMD PMD Saltatory (jumping) nerve conduction Investigating the Myelinogenic Potential of Individual Oligodendrocytes In Vivo Sparse Labeling of Oligodendrocytes CNPase-GFP Variegated expression under the MBP-enhancer Cerebral Cortex Corpus Callosum Cerebral Cortex Corpus Callosum Cerebral Cortex Caudate Putamen Corpus Callosum Cerebral Cortex Caudate Putamen Corpus Callosum Corpus Callosum Cerebral Cortex Caudate Putamen Corpus Callosum Ant Commissure Corpus Callosum Cerebral Cortex Caudate Putamen Piriform Cortex Corpus Callosum Ant Commissure Characterization of Oligodendrocyte Morphology Cerebral Cortex Corpus Callosum Caudate Putamen Cerebellum Brain Stem Spinal Cord Oligodendrocytes in disease: Cerebral Palsy ! CP major cause of chronic neurological morbidity and mortality in children ! CP incidence now about 3/1000 live births compared to 1/1000 in 1980 when we started intervening for ELBW ! Of all ELBW {gestation 6 mo, Wt. 0.5kg} , 10-15% develop CP ! Prematurely born children prone to white matter injury {WMI}, principle reason for the increase in incidence of CP ! ! 12 Cerebral Palsy Spectrum of white matter injury ! ! Macro Cystic Micro Cystic Gliotic Khwaja and Volpe 2009 13 Rationale for Repair/Remyelination in Multiple Sclerosis Oligodendrocyte specification oligodendrocytes specified from the pMN after MNs - a ventral source of oligodendrocytes -
11 Introduction to the Nervous System and Nervous Tissue
11 Introduction to the Nervous System and Nervous Tissue ou can’t turn on the television or radio, much less go online, without seeing some- 11.1 Overview of the Nervous thing to remind you of the nervous system. From advertisements for medications System 381 Yto treat depression and other psychiatric conditions to stories about celebrities and 11.2 Nervous Tissue 384 their battles with illegal drugs, information about the nervous system is everywhere in 11.3 Electrophysiology our popular culture. And there is good reason for this—the nervous system controls our of Neurons 393 perception and experience of the world. In addition, it directs voluntary movement, and 11.4 Neuronal Synapses 406 is the seat of our consciousness, personality, and learning and memory. Along with the 11.5 Neurotransmitters 413 endocrine system, the nervous system regulates many aspects of homeostasis, including 11.6 Functional Groups respiratory rate, blood pressure, body temperature, the sleep/wake cycle, and blood pH. of Neurons 417 In this chapter we introduce the multitasking nervous system and its basic functions and divisions. We then examine the structure and physiology of the main tissue of the nervous system: nervous tissue. As you read, notice that many of the same principles you discovered in the muscle tissue chapter (see Chapter 10) apply here as well. MODULE 11.1 Overview of the Nervous System Learning Outcomes 1. Describe the major functions of the nervous system. 2. Describe the structures and basic functions of each organ of the central and peripheral nervous systems. 3. Explain the major differences between the two functional divisions of the peripheral nervous system. -

How Does the Brain Work? Grades 9-12
Fact Sheet For Grades 9-12 When a neuron is at rest, the cell body, or How does the soma, of the neuron is negatively charged relative to the outside of the neuron. A neuron at rest has a brain work? negative charge of approximately -70 millivolts By Elizabeth A. Weaver II and Hillary H. Doyle (mV) of electricity. However, when a stimulus comes along (like stubbing your toe, or hearing your name being called), it causes the neuron to With 80-100 billion nerve cells, known as take in more positive ions, and the neuron neurons, the human brain is capable of some becomes more positively charged. Once the astonishing feats. Each neuron is connected to neuron reaches a certain threshold of more than 1,000 other neurons, making the total approximately -55mV, an event known as an action number of connections in the brain around 60 potential occurs and causes the neuron to “fire”. trillion! Neurons are organized into patterns and The action potential travels down the axon where networks within the brain and communicate with it reaches the axon terminal. each other at incredible speeds. At the axon terminal, electrical signals are Each neuron is made up of three main converted into chemical signals that travel parts: the cell body (also known as the soma), the between neurons across a small gap called the axon, and the dendrites. Neurons communicate synapse. These chemicals are called with each other using electrochemical signals. In neurotransmitters. Neurotransmitters cross the other words, certain chemicals in the body known synapse and attach to receptors on the dendrites as ions have an electrical charge. -

Intersegmental Interneurons Can Control the Gain of Reflexes in Adjacent Segments of the Locust by Their Action on Nonspiking Local Interneurons
The Journal of Neuroscience, September 1989, g(9): 30303039 Intersegmental Interneurons Can Control the Gain of Reflexes in Adjacent Segments of the Locust by Their Action on Nonspiking Local Interneurons Gilles Laurent and Malcolm Burrows Department of Zoology, University of Cambridge, Cambridge CB2 3EJ, England The gain of local reflexes of one leg of a locust can be altered partmentalized, with synaptic inputs and their associated by mechanosensory inputs generated by movements of or conductance changes restricted to particular branches. In tactile inputs to an adjacent leg. Touching the mesothoracic this way, an individual nonspiking neuron could contribute tarsus, for example, increases the number of spikes that are simultaneously to several local circuits. The inputs from dif- produced by the metathoracic slow extensor tibiae motor ferent intersegmental interneurons could then modulate these neuron and enhances the depolarization of flexor tibiae mo- pathways independently. tor neuron in response to imposed movements of the chor- dotonal organ in the ipsilateral hind femur. The sensory in- Nonspiking interneurons in the metathoracic ganglion of the formation from the middle leg is conveyed directly to locust receive direct inputs from mechanosensory afferents on nonspiking interneurons and motor neurons controlling the one hindleg and are essential elements in local reflex movements movements of the hindleg by a population of mesothoracic of that leg (Laurent and Burrows, 1988; Burrows et al., 1988). intersegmental interneurons (Laurent and Burrows, 1989). They also receive direct inputs from intersegmental intemeu- The metathoracic nonspiking interneurons receive direct in- rons that process the mechanosensory inputs from an ipsilateral puts from receptors on a hindleg and are, therefore, a point middle leg (Laurent and Burrows, 1989). -

Neuron-Satellite Glial Cell Interactions in Sympathetic Nervous System Development
NEURON-SATELLITE GLIAL CELL INTERACTIONS IN SYMPATHETIC NERVOUS SYSTEM DEVELOPMENT by Erica D. Boehm A dissertation submitted to the Johns Hopkins University in conformity with the requirements for the degree of Doctor of Philosophy Baltimore, Maryland July 2020 © 2020 Erica Boehm All rights reserved. ABSTRACT Glial cells play crucial roles in maintaining the stability and structure of the nervous system. Satellite glial cells are a loosely defined population of glial cells that ensheathe neuronal cell bodies, dendrites, and synapses of the peripheral nervous system (Elfvin and Forsman 1978; Pannese 1981). Satellite glial cells are closely juxtaposed to peripheral neurons with only 20nm of space between their membranes (Dixon 1969). This close association suggests a tight coupling between the cells to allow for possible exchange of important nutrients, yet very little is known about satellite glial cell function and development. How neurons and glial cells co-develop to create this tightly knit unit remains undefined, as well as the functional consequences of disrupting these contacts. Satellite glial cells are derived from the same population of cells that give rise to peripheral neurons, but do not begin differentiation and proliferation until neurogenesis has been completed (Hall and Landis 1992). A key signaling pathway involved in glial specification is the Delta/Notch signaling pathway (Tsarovina et al. 2008). However, recent studies also implicate Notch signaling in the maturation of glia through non- canonical Notch ligands such as Delta/Notch-like EGF-related Receptor (DNER) (Eiraku et al. 2005). Interestingly, it has been reported that levels of DNER in sympathetic neurons may be dependent on the target-derived growth factor, nerve growth factor (NGF), and this signal is prominent in sympathetic neurons at the time in which satellite glial cells are developing (Deppmann et al. -

Normal Myelination a Practical Pictorial Review
Normal Myelination A Practical Pictorial Review Helen M. Branson, BSc, MBBS, FRACR KEYWORDS Myelin Myelination T1 T2 MR Diffusion KEY POINTS MR imaging is the best noninvasive modality to assess myelin maturation in the human brain. A combination of conventional T1-weighted and T2-weighted sequences is all that is required for basic assessment of myelination in the central nervous system (CNS). It is vital to have an understanding of the normal progression of myelination on MR imaging to enable the diagnosis of childhood diseases including leukodystrophies as well as hypomyelinating disorders, delayed myelination, and acquired demyelinating disease. INTRODUCTION and its role in the human nervous system is needed. Assessment of the progression of myelin and mye- Myelin is present in both the CNS and the lination has been revolutionized in the era of MR peripheral nervous system. In the CNS, it is imaging. Earlier imaging modalities such as ultra- primarily found in white matter (although small sonography and computed tomography have no amounts are also found in gray matter) and thus current role or ability to contribute to the as- is responsible for its color.1 Myelin acts as an elec- sessment of myelin maturation or abnormalities trical insulator for neurons.1 Myelin plays a role in of myelin. The degree of brain myelination can be increasing the speed of an action potential by used as a marker of maturation. 10–100 times that of an unmyelinated axon1 and The authors discuss also helps in speedy axonal transport.2 Edgar 3 1. Myelin function and structure and Garbern (2004) demonstrated that the ab- 2. -

Neurotransmission Fact Sheet
Neurotransmission Fact Sheet The brain and nervous system are made of billions of nerve cells, called neurons. Neurons have three main parts: cell body, dendrites, and axon. The axon is covered by the myelin sheath. Cell Body is in charge of the neuron’s Dendrites receive messages activities. from other neurons. Myelin Sheath covers the axon to protect it and help messages travel faster and easier. Axon sends messages from the cell body to the dendrites of other neurons. The transfer of information between neurons is called neurotransmission. This is how neurotransmission works: 1. A message travels from the dendrites through the cell body and to the end of the axon. 2. The message causes the chemicals, called neurotransmitters, to be released from the end of the axon into the synapse. The neurotransmitters carry the message with them into the synapse. The synapse is the space between the axon of one neuron and the dendrites of another neuron. 3. The neurotransmitters then travel across the synapse to special places on the dendrites of the next neuron, called receptors. The neurotransmitters fit into the receptors like keys in locks. 4. Once the neurotransmitter has attached to the receptors of the second neuron, the message is passed on. 5. The neurotransmitters are released from the receptors and are either broken down or go back into the axon of the first neuron. Teacher copy: Module 1 Neurotransmission Fact Sheet The brain and nervous system are made of billions of nerve cells, called neurons. Neurons have three main parts: cell body, dendrites, and axon. -

NERVE TISSUE Neuron – Nerve Cell
Department of Histology and Embryology, P. J. Šafárik University, Medical Faculty, Košice NERVE TISSUE: Sylabus for foreign students Author: doc. MVDr. Iveta Domoráková, PhD. Revised by: prof. MUDr. Eva Mechírová, CSc. NERVE TISSUE FUNCTION: Reception, transmission, processing of nerve stimuli. Coordination of all functional activities in the body: - motor function (body movement) - sensory (rapid response to external stimuli) - visceral, endocrine and exocrine glands - mental functions, memory, emotion A) Anatomically nervous system consists of: 1. CNS (central nervous system) – brain, spinal cord 2. PNS (peripheral nervous system) – peripheral nerves and ganglia B) Functionally nervous system is divided into the: 1. Somatic nervous system (sensory and motor innervation) 2. Autonomic nervous system (involuntary innervation of smooth muscles, glands) C) Microscopic structure of the nerve tissue - two types of cells: 1. Nerve cells – neurons 2. Glial cells (supporting, electrical insulation, metabolic function) Neuron – nerve cell - is the structural and functional unit of the nerve tissue - receives stimuli from other cells - conducts electrical impulses to another cells by their processes - chainlike communication - ten bilion of neurons in humans A. Neurons according the shape: Pyramidal (E) star-shaped (D) pear-shaped (G) oval (B) B. Types of neurons according number of the processes 1. multipolar (D,E,G)) 2. bipolar (A) 3. pseudounipolar (B) 4. unipolar C. Neurons - according the function Motor (efferent) neurons – convey impulses -

Evidence for Oligodendrocyte Progenitor Cell Heterogeneity in the Adult Mouse Brain
bioRxiv preprint doi: https://doi.org/10.1101/2020.03.06.981373; this version posted March 8, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Evidence for oligodendrocyte progenitor cell heterogeneity in the adult mouse brain. Authors: Rebecca M. Beiter1,2, Anthony Fernández-Castañeda1,2, Courtney Rivet-Noor1,2, Andrea Merchak1,2, Robin Bai1, Erica Slogar1, Scott M. Seki1,2,3, Dorian A Rosen1,4, Christopher C. Overall1,5, and Alban Gaultier1,5,# 1Center for Brain Immunology and Glia, Department of Neuroscience, 2Graduate Program in Neuroscience, 3Medical Scientist Training Program, 4Graduate Program in Pharmacological Sciences School of Medicine, University of Virginia, Charlottesville, VA 22908, USA. #Corresponding author. Email: [email protected] 5Christopher C. Overall and Alban Gaultier are co-senior authors 1 bioRxiv preprint doi: https://doi.org/10.1101/2020.03.06.981373; this version posted March 8, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. ABSTRACT Oligodendrocyte progenitor cells (OPCs) are a mitotically active population of glia that comprise approximately 5% of the adult brain. OPCs maintain their proliferative capacity and ability to differentiate in oligodendrocytes throughout adulthood, but relatively few mature oligodendrocytes are produced following developmental myelination. Recent work has begun to demonstrate that OPCs likely perform multiple functions in both homeostasis and disease, and can significantly impact behavioral phenotypes such as food intake and depressive symptoms. However, the exact mechanisms through which OPCs might influence brain function remains unclear.