A New Functional JAZ Degron Sequence in Strawberry JAZ1
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Rhodotypos Scandens Common Name: Black Jetbead Family Name
Plant Profiles: HORT 2241 Landscape Plants I Botanical Name: Rhodotypos scandens Common Name: black jetbead Family Name: Rosaceae –rose family General Description: Rhodotypos scandens is a tough, adaptable flowering shrub. It has white flowers in late spring, handsome leaves during the summer and fall, and interesting small black fruits that hold on during the winter. It does well in sun or dense shade and is tolerant of a wide variety of landscape conditions. Rhodotypos scandens was introduced from Asia for use as an ornamental plant but has become an invasive species in eastern United States. Though not a widespread problem in this area, it has been documented in natural areas in DuPage, Cook and a few other areas on the Chicago area. Zone: 4-8 Resources Consulted: Dirr, Michael A. Manual of Woody Landscape Plants: Their Identification, Ornamental Characteristics, Culture, Propagation and Uses. Champaign: Stipes, 2009. Print. "The PLANTS Database." USDA, NRCS. National Plant Data Team, Greensboro, NC 27401-4901 USA, 2014. Web. 23 Mar. 2014. Swink, Floyd, and Gerould Wilhelm. Plants of the Chicago Region. Indianapolis: Indiana Academy of Science, 1994. Print. Creator: Julia Fitzpatrick-Cooper, Professor, College of DuPage Creation Date: 2014 Keywords/Tags: Rhodotypos scandens, deciduous, flowering shrub, shrub Whole plant/Habit: Description: Rhodotypos scandens has an upright, arching habit that resembles a Japanese kerria (Kerria japonica) on steroids! The loose arching stems grow 3-6 foot tall and 6-9 foot wide. Image Source: Karren Wcisel, TreeTopics.com Image Date: May 6, 2005 Image File Name: jetbead_0529.png Flower: Description: The four-petaled white flowers bloom mid-spring to early summer. -

Staff Summary for April 15-16, 2020
Item No. 30 STAFF SUMMARY FOR APRIL 15-16, 2020 30. SHASTA SNOW-WREATH CESA PETITION Today’s Item Information ☐ Action ☒ Consider and potentially act on the petition, DFW’s evaluation report, and comments received to determine whether listing Shasta snow-wreath (Neviusia cliftonii) as a threatened or endangered species under the California Endangered Species Act (CESA) may be warranted. Summary of Previous/Future Actions • Received petition Sep 30, 2019 • FGC transmitted petition to DFW Oct 10, 2019 • Published notice of receipt of petition Nov 22, 2020 • Public receipt of petition Dec 11-12, 2019; Sacramento • Received DFW 90-day evaluation report Feb 21, 2020; Sacramento • Today, determine if petitioned action Apr 15-16, 2020; Teleconference may be warranted Background A petition to list Shasta snow-wreath as endangered under CESA was submitted by Kathleen Roche and the California Native Plant Society on Sep 30, 2019 (Exhibit 1). On Oct 10, 2019, FGC staff transmitted the petition to DFW for review. A notice of receipt of petition was published in the California Regulatory Notice Register on Nov 22, 2019. California Fish and Game Code Section 2073.5 requires that DFW evaluate the petition and submit to FGC a written evaluation with a recommendation, which was received at FGC’s Feb 21, 2020 meeting. The evaluation report (Exhibit 2) delineates each of the categories of information required for a petition, evaluates the sufficiency of the available scientific information for each of the required components, and incorporates additional relevant information that DFW possessed or received during the review period. Today’s agenda item follows the public release and review period of the evaluation report prior to FGC action, as required in Fish and Game Code Section 2074. -

Kerria Japonica (Kerria) a Deciduous Shrub That Blooms Yellow Flowers in Spring
Kerria japonica (kerria) A deciduous shrub that blooms yellow flowers in spring. It thrives in full sun and well drained soil but after it gets established it can tolerate drought to a certain extent. It is in best shape when given space to spread because of its arching habit Landscape Information Pronounciation: KAIR-ee-uh juh-PON-ih-kuh Plant Type: Shrub Origin: South Asia, India Heat Zones: 1, 2, 3, 4, 5, 6, 7, 8, 9 Hardiness Zones: 5, 6, 7, 9 Uses: Hedge, Specimen, Border Plant, Cut Flowers / Arrangements Size/Shape Growth Rate: Fast Tree Shape: Round, Spreading, Vase Canopy Texture: Medium Height at Maturity: 1.5 to 3 m Plant Image Spread at Maturity: 1.5 to 3 meters Time to Ultimate Height: 5 to 10 Years Kerria japonica (kerria) Botanical Description Foliage Leaf Arrangement: Alternate Leaf Venation: Pinnate Leaf Persistance: Deciduous Leaf Type: Simple Leaf Blade: 5 - 10 cm Leaf Shape: Lanceolate Leaf Margins: Serrulate, Double Serrate Leaf Scent: Color(growing season): Green Color(changing season): Yellow Flower Flower Showiness: True Flower Type: Solitary Flower Image Flower Scent: Pleasant Flower Color: Yellow Seasons: Spring Trunk Trunk Esthetic Values: Showy, Smooth Fruit Fruit Type: Follicle Fruit Showiness: False Fruit Colors: Brown, Red Seasons: Summer Kerria japonica (kerria) Horticulture Management Tolerance Drought Tolerant: Yes Requirements Soil Requirements: Clay, Loam, Sand Soil Ph Requirements: Acidic, Neutral, Alkaline Water Requirements: Moderate Light Requirements: Full, Part Management Edible Parts: Plant Propagations: Cutting, Division Leaf Image MORE IMAGES Other Image. -

Pruning and Training Small Trees and Shrubs
Pruning and Training Average Life Expectancy of Small Trees and Shrubs Street Trees Downtown: 7 years City Average: 32 years Best City Site: 60 years Rural Site: 150 years Dr. Laura G. Jull Dept. of Horticulture University of Wisconsin-Madison Plant Selection Prune based on client’s One of the most important decisions needs but keep in mind Poor plant choices become plant health and natural maintenance problems form of plant Plant the right plant in the right place Use adaptable plants specific to site, whether native or not See UW-Extension Publication A- 3864: Choosing the Right Landscape Plants: Factors to Consider at http://learningstore.uwex.edu Objectives of Pruning Principles of Good Pruning Control size Always assess entire plant before making any cuts Direct and train growth Never prune without a good Influence flowering and fruiting reason Corrective pruning Keep in mind growth rate of Maintain health and appearance plant, form, mature height and . Remove dead, diseased, or damaged tissue width Safety: hazard trees Don’t forget flower and fruit Rejuvenate old, overgrown plants display Specialty pruning: topiary, espalier, All plants have a finite life, pleaching, pollarding, bonsai may not be worth rejuvenating half dead shrub 1 General Responses to Pruning General Responses to Pruning Responds by making new Breaks apical dominance growth elsewhere Removal of terminal shoot/bud Stimulates budbreak at cut . Sap flow stops to terminal Response varies based on: . Shoots just below wound sprout . Growth habit Related to reduced flow of auxin . Plant age Common example: pinching of tips of annuals causes bushiness . Size Removal of lateral shoot . -

IHCA Recommended Plant List
Residential Architectural Review Committee Recommended Plant List Plant Materials The following plant materials are intended to guide tree and shrub ADDITIONS to residential landscapes at Issaquah Highlands. Lot sizes, shade, wind and other factors place size and growth constraints on plants, especially trees, which are suitable for addition to existing landscapes. Other plant materials may be considered that have these characteristics and similar maintenance requirements. Additional species and varieties may be selected if authorized by the Issaquah Highlands Architectural Review Committee. This list is not exhaustive but does cover most of the “good doers” for Issaquah Highlands. Our microclimate is colder and harsher than those closer to Puget Sound. Plants not listed should be used with caution if their performance has not been observed at Issaquah Highlands. * Drought-tolerant plant ** Requires well-drained soil DECIDUOUS TREES: Small • Acer circinatum – Vine Maple • Acer griseum – Paperbark Maple • *Acer ginnala – Amur Maple • Oxydendrum arboreum – Sourwood • Acer palmation – Japanese Maple • *Prunus cerasifera var. – Purple Leaf Plum varieties • Amelanchier var. – Serviceberry varieties • Styrax japonicus – Japanese Snowbell • Cornus species, esp. kousa Medium • Acer rufinerve – Redvein Maple • Cornus florida (flowering dogwood) • *Acer pseudoplatanus – Sycamore Maple • Acer palmatum (Japanese maple, many) • • *Carpinus betulus – European Hornbeam Stewartia species (several) • *Parrotia persica – Persian Parrotia Columnar Narrow -

Kerria Japonica
Woody Plants Database [http://woodyplants.cals.cornell.edu] Species: Kerria japonica (kare'ee-ah ja-pon'i-kah) Japanese Kerria; Kerria Rose Cultivar Information * See specific cultivar notes on next page. Ornamental Characteristics Size: Shrub 4 to 8 feet Height: 3' - 6' (spread 9') Leaves: Deciduous Shape: rounded Ornamental Other: full sun to part shade Environmental Characteristics Light: Part shade Hardy To Zone: 5b Soil Ph: Can tolerate acid to alkaline soil (pH 5.0 to 8.0) Environmental Other: full sun to part shade Insect Disease trouble free Bare Root Transplanting Moderately difficult Other native to China; transplant B & B or container Moisture Tolerance 1 Woody Plants Database [http://woodyplants.cals.cornell.edu] Occasionally saturated Consistently moist, Occasional periods of Prolonged periods of or very wet soil well-drained soil dry soil dry soil 1 2 3 4 5 6 7 8 9 10 11 12 2 Woody Plants Database [http://woodyplants.cals.cornell.edu] Cultivars for Kerria japonica Showing 1-7 of 7 items. Cultivar Name Notes Pleniflora 'Pleniflora' - double flowers Variegata 'Variegata' (a.k.a. 'Picta') - leaves edged in white Golden Guinea 'Golden Guinea' - large flowers (to 2" wide); long blooming period Kin Kan 'Kin Kan' (a.k.a. 'Aureovittata') - stems color up yellow with thin green stripes in winter; frequently reverts Shannon 'Shannon' - vigorous; grows to 6' tall; blooms earlier than species Chiba Gold ‘Chiba Gold’ - gold to chartreuse foliage, colors well in shade Honshu ‘Honshu’ - larger, 2-inch, single flowers with petals that overlap -

Annual Benefit Plant Sale 2012
Annual Benefit Plant Sale 2012 Botanic Gardens COLLEGE OF AGRICULTURE & NATURAL RESOURCES Connect to nature Get inspired by wildflowers, naturalistic gardening and meadows in a whole new way with our seasonal garden tours. Enjoy an art class in the garden or learn about native plant gardening, conservation, and habitats by taking one of our classes. And if you can’t visit us, enroll in our new online distance learning program, Mt. Cuba Center Connect. Visit www.mtcubacenter.org to reserve a tour or sign up for a class. Two-Hour Guided Tours | $5 per person Spring Wildflower Tours April 12th – May 27th Summer Twilight Tours May 30th – July 26th 8th Annual Wildflower Celebration |Free th April 29 , 10am – 4pm Purple pitcherplant (Sarracenia purpurea) Greenville, DE P: 302.239.4244 www.mtcubacenter.org INSPIRATION x EDUCATION x CONSERVATION 2 2012 SPRING PLANT SALE CATALOG WEBSITE: http://ag.udel.edu/udbg/events/annualsale.html WELCOME We welcome you to the twentieth annual UDBG benefit plant sale. In addition to its role as the major source of funding for the UDBG, 2012 BENEFIT PLANT SALE CATALOG we hope it also serves as a major educational event for our members and the public. It presents an opportunity to learn about new plants and consider possibilities. We should always look for ways to expand and improve our knowledge about plants and this catalog offers possibilities to accomplish both. As always, we offer an in- depth look at a particular group of plants, this year the genus Camellia. The selection goes beyond offering various cultivars with differing flower color, to a more extensive exploration of the genus with particular emphasis on hardy selections and new hybrids Camellia ‘Autumn Spirit’. -

Don't Plant a Pest!
Many of the characteristics that make a plant Gardening green an attractive choice for the garden may also make it a successful invader: California is a gardener’s dream. Our mild climate Don’t allows us to have fantastic gardens, showcasing a wide Garden Plants: Invasive Plants: variety of ornamental Easy to propagate Broad germination plants from all around Establish rapidly Colonizer the world. Mature early Mature early Abundant flowers Prolific seeds plant a But sometimes, our Pest/disease tolerant Few natural predators garden plants “jump the fence” and invade Invasive plants are by nature a regional problem. A natural areas. These plant that jumps out of the garden in one climate and invasive plants can habitat type may behave perfectly in another. The become serious wildland twelve problem plants listed here have escaped from pest! weeds that threaten gardens throughout the greater Bay Area. California’s biodiversity and economy. How to use this brochure: This brochure suggests safe alternatives for these More than half of the plants. When you are buying new plants, consider these Give them an inch and plants currently Conservancy Nature The Rice, Barry alternatives, or ask your local nursery for other damaging California’s French broom invades Bay Area hillsides non-invasive plants. If one of the invasive plants is wildlands were originally introduced for landscaping already in your yard, especially if you live near they’ll take an acre... purposes. Garden escapes like pampasgrass and Scotch wildlands, you may want to remove it and replace it broom may have desirable characteristics in a garden with a recommended alternative. -

Trees, Shrubs, and Perennials That Intrigue Me (Gymnosperms First
Big-picture, evolutionary view of trees and shrubs (and a few of my favorite herbaceous perennials), ver. 2007-11-04 Descriptions of the trees and shrubs taken (stolen!!!) from online sources, from my own observations in and around Greenwood Lake, NY, and from these books: • Dirr’s Hardy Trees and Shrubs, Michael A. Dirr, Timber Press, © 1997 • Trees of North America (Golden field guide), C. Frank Brockman, St. Martin’s Press, © 2001 • Smithsonian Handbooks, Trees, Allen J. Coombes, Dorling Kindersley, © 2002 • Native Trees for North American Landscapes, Guy Sternberg with Jim Wilson, Timber Press, © 2004 • Complete Trees, Shrubs, and Hedges, Jacqueline Hériteau, © 2006 They are generally listed from most ancient to most recently evolved. (I’m not sure if this is true for the rosids and asterids, starting on page 30. I just listed them in the same order as Angiosperm Phylogeny Group II.) This document started out as my personal landscaping plan and morphed into something almost unwieldy and phantasmagorical. Key to symbols and colored text: Checkboxes indicate species and/or cultivars that I want. Checkmarks indicate those that I have (or that one of my neighbors has). Text in blue indicates shrub or hedge. (Unfinished task – there is no text in blue other than this text right here.) Text in red indicates that the species or cultivar is undesirable: • Out of range climatically (either wrong zone, or won’t do well because of differences in moisture or seasons, even though it is in the “right” zone). • Will grow too tall or wide and simply won’t fit well on my property. -
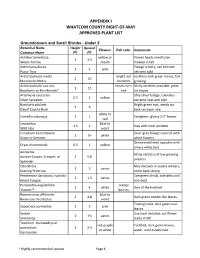
Whatcom County's Approved Plant List
APPENDIX I WHATCOM COUNTY RIGHT-OF-WAY APPROVED PLANT LIST Groundcovers and Small Shrubs - Under 2' Botanical Name Height Spread Flowers Fall color Comments Common Name (ft) (ft) Achillea tomentosa yellow or Flower heads need to be 1 3-5 Wooly Yarrow cream mowed in fall Antennaria dioica Foliage is furry, can tolerate 1 2 pink Pussy Toes extreme cold Arctostaphylos media bright red Leathery dark green leaves, fast 2 10 Manzanita Media branches growing Arctostaphylos uva ursi leaves turn Many varieties available, great 1 15 Bearberry or Kinnikinnick* red for slopes Artemesia caucasica Silky silver foliage, tolerates 0.5 2 yellow Silver Spreader extreme heat and cold Baccharis pilularis Bright green mat, needs cut 2 6 Dwarf Coyote Bush back once per year white to Camellia sasanqua 1 1 Evergreen, glossy 2-3" leaves red Ceanothus blue to 1.5 2 Stay with local varieties Wild Lilac violet Cerastium tomentosum Silver gray foliage covered with 1 5+ white Snow-in-Summer white flowers Ornamental seed capsules with Dryas drummondii 0.5 1 yellow silvery white tails Juniperus Many varieties of low growing Juniper-Carpet, Creeper, or 2 5-8 junipers Spreader Oenothera May die back in severe winters, 1 2 varies Evening Primrose come back strong Penstemon davidsonii, rupicola Evergreen shrub, tolerates cold 2 1.5 varies Beard Tongue and heat Pyracantha augustifolia orange 1 4 white One of the hardiest 'Gnome'* berries Rosmariunus officinalis blue to 2 4-8 Dark green needle-like leaves Rosemary 'Prostratus' violet Trailing habit, dark green oval Saponaria ocymoides 1 3 pink leaves Sedum Use local varieties, cut flower 2 1-5 varies Stonecrop stalks in fall Teucrium chamaedrys or red-purple Toothed, dark green leaves, postratium 1 2-3 or white water until established Germander * Highly-recommended species Page 1 Thymus white to 2 1-2 Many varieties Thyme purple Shrubs Botanical Name Height Spread Flowers Fall color Comments Common Name (ft) (ft) Arctostaphylos bright red Shiny dark green leaves turn 2-15 20 varies Manzanita berries maroon in fall. -

Oakland Nurseries Inc Japanese Kerria
Japanese Kerria Kerria japonica Height: 6 feet Spread: 6 feet Sunlight: Hardiness Zone: 4b Description: Japanese Kerria flowers A colorful spring blooming shrub featuring bright golden Photo courtesy of NetPS Plant Finder flowers on thin, colorful stems and light green leaves; spreads by suckering to form a natural mass; use in groupings in partial shade in the garden, complemented by other plants Ornamental Features Japanese Kerria features showy yellow flowers along the branches from early to mid spring. It has light green foliage throughout the season. The narrow leaves turn lemon yellow in fall. The fruit is not ornamentally significant. The smooth bark and lime green branches add an interesting dimension to the landscape. Landscape Attributes Japanese Kerria in bloom Photo courtesy of NetPS Plant Finder Japanese Kerria is a dense multi-stemmed deciduous shrub with an upright spreading habit of growth. It lends an extremely fine and delicate texture to the landscape composition which can make it a great accent feature on this basis alone. This is a high maintenance shrub that will require regular care and upkeep, and should only be pruned after flowering to avoid removing any of the current season's flowers. Deer don't particularly care for this plant and will usually leave it alone in favor of tastier treats. Gardeners should be aware of the following characteristic(s) that may warrant special consideration; - Suckering Japanese Kerria is recommended for the following landscape applications; - Mass Planting - General Garden Use Columbus -

Guidebook to Invasive Nonnative Plants of the Elwha Watershed Restoration
Guidebook to Invasive Nonnative Plants of the Elwha Watershed Restoration Olympic National Park, Washington Cynthia Lee Riskin A project submitted in partial fulfillment of the requirements for the degree of Master of Environmental Horticulture University of Washington 2013 Committee: Linda Chalker-Scott Kern Ewing Sarah Reichard Joshua Chenoweth Program Authorized to Offer Degree: School of Environmental and Forest Sciences Guidebook to Invasive Nonnative Plants of the Elwha Watershed Restoration Olympic National Park, Washington Cynthia Lee Riskin Master of Environmental Horticulture candidate School of Environmental and Forest Sciences University of Washington, Seattle September 3, 2013 Contents Figures ................................................................................................................................................................. ii Tables ................................................................................................................................................................. vi Acknowledgements ....................................................................................................................................... vii Introduction ....................................................................................................................................................... 1 Bromus tectorum L. (BROTEC) ..................................................................................................................... 19 Cirsium arvense (L.) Scop. (CIRARV)