Genetic Diversity and Differentiation of Invasive Acacia Longifolia in Portugal
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
Additional Information
Current Survey Introduced Flora Records Vegetation Condition *Acetosa vesicaria Excellent 534,000 mE 534,000 mE 535,000 534,000 mE 534,000 mE 535,000 534,000 mE 534,000 mE 535,000 534,000 mE 534,000 mE 535,000 534,000 mE 534,000 mE 535,000 534,000 mE 534,000 mE 535,000 534,000 mE 534,000 mE 535,000 536,000 mE 536,000 537,000 mE 537,000 536,000 mE 536,000 537,000 mE 537,000 536,000 mE 536,000 537,000 mE 537,000 536,000 mE 536,000 537,000 mE 537,000 536,000 mE 536,000 537,000 mE 537,000 536,000 mE 536,000 537,000 mE 537,000 536,000 mE 536,000 537,000 mE 537,000 534,000 mE 534,000 mE 535,000 534,000 mE 534,000 mE 535,000 534,000 mE 534,000 mE 535,000 534,000 mE 534,000 mE 535,000 534,000 mE 534,000 mE 535,000 534,000 mE 534,000 mE 535,000 534,000 mE 534,000 mE 535,000 536,000 mE 536,000 537,000 mE 537,000 536,000 mE 536,000 537,000 mE 537,000 536,000 mE 536,000 537,000 mE 537,000 536,000 mE 536,000 537,000 mE 537,000 536,000 mE 536,000 537,000 mE 537,000 536,000 mE 536,000 537,000 mE 537,000 536,000 mE 536,000 537,000 mE 537,000 534,000 mE 534,000 mE 535,000 534,000 mE 534,000 mE 535,000 534,000 mE 534,000 mE 535,000 534,000 mE 534,000 mE 535,000 534,000 mE 534,000 mE 535,000 534,000 mE 534,000 mE 535,000 534,000 mE 534,000 mE 535,000 536,000 mE 536,000 537,000 mE 537,000 536,000 mE 536,000 537,000 mE 537,000 536,000 mE 536,000 537,000 mE 537,000 536,000 mE 536,000 537,000 mE 537,000 536,000 mE 536,000 537,000 mE 537,000 536,000 mE 536,000 537,000 mE 537,000 536,000 mE 536,000 537,000 mE 537,000 534,000 mE 534,000 mE 535,000 534,000 mE 534,000 -
(A) Journals with the Largest Number of Papers Reporting Estimates Of
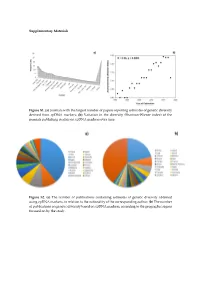
Supplementary Materials Figure S1. (a) Journals with the largest number of papers reporting estimates of genetic diversity derived from cpDNA markers; (b) Variation in the diversity (Shannon-Wiener index) of the journals publishing studies on cpDNA markers over time. Figure S2. (a) The number of publications containing estimates of genetic diversity obtained using cpDNA markers, in relation to the nationality of the corresponding author; (b) The number of publications on genetic diversity based on cpDNA markers, according to the geographic region focused on by the study. Figure S3. Classification of the angiosperm species investigated in the papers that analyzed genetic diversity using cpDNA markers: (a) Life mode; (b) Habitat specialization; (c) Geographic distribution; (d) Reproductive cycle; (e) Type of flower, and (f) Type of pollinator. Table S1. Plant species identified in the publications containing estimates of genetic diversity obtained from the use of cpDNA sequences as molecular markers. Group Family Species Algae Gigartinaceae Mazzaella laminarioides Angiospermae Typhaceae Typha laxmannii Angiospermae Typhaceae Typha orientalis Angiospermae Typhaceae Typha angustifolia Angiospermae Typhaceae Typha latifolia Angiospermae Araliaceae Eleutherococcus sessiliflowerus Angiospermae Polygonaceae Atraphaxis bracteata Angiospermae Plumbaginaceae Armeria pungens Angiospermae Aristolochiaceae Aristolochia kaempferi Angiospermae Polygonaceae Atraphaxis compacta Angiospermae Apocynaceae Lagochilus macrodontus Angiospermae Polygonaceae Atraphaxis -

A Vegetation and Flora Survey of the Brockman Syncline 4 Project Area, Near Tom Price
AA VVeeggeettaattiioonn aanndd FFlloorraa SSuurrvveeyy ooff tthhee BBrroocckkmmaann SSyynncclliinnee 44 PPrroojjeecctt AArreeaa,, nneeaarr TToomm PPrriiccee Prepared for Hamersley Iron Pty Ltd Prepared by JJuulllyy 22000055 Biota Environmental Sciences Pty Ltd A Vegetation and Flora Survey of the Brockman Syncline 4 Project Area, near Tom Price © Biota Environmental Sciences Pty Ltd 2005 ABN 49 092 687 119 14 View Street North Perth Western Australia 6006 Ph: (08) 9328 1900 Fax: (08) 9328 6138 Project No.: 271 Prepared by: Michi Maier Checked by: Garth Humphreys This document has been prepared to the requirements of the client identified on the cover page and no representation is made to any third party. It may be cited for the purposes of scientific research or other fair use, but it may not be reproduced or distributed to any third party by any physical or electronic means without the express permission of the client for whom it was prepared or Biota Environmental Sciences Pty Ltd. Cube:Current:271 (Brockman 4 Biological):Doc:flora:flora_survey_7.doc 2 A Vegetation and Flora Survey of the Brockman Syncline 4 Project Area, near Tom Price A Vegetation and Flora Survey of the Brockman Syncline 4 Project Area, near Tom Price Contents 1.0 Summary 6 1.1 Background 6 1.2 Vegetation 6 1.3 Flora 7 1.4 Management Recommendations 7 2.0 Introduction 9 2.1 Background to the BS4 Project and Location of the Project Area 9 2.2 Scope and Objectives of this Study 9 2.3 Purpose of this Report 12 2.4 Existing Environment 12 3.0 Methodology 18 3.1 Desktop -

SCIENCE COMMUNICATIONS No
SCIENCE COMMUNICATIONS No. 34 (January 2014) Science & Conservation Division Mission: The provision of up-to-date and scientifically sound information to uphold effective evidence-based conservation of biodiversity and sustainable natural resource management in Western Australia Science Communications lists titles of Davison EM, McGurk LE, Bougher NL, Syme K, publications, reports etc prepared by Science Watkin ELJ (2013). Amanita lesueuri and and Conservation Division staff. This issue is for Amanita wadjukorum (Basidiomycota), two new the period August 2013.-January 2014 species from Western Australia, and an If you require a copy of a submitted paper please expanded description of Amanita fibrillopes. contact the author. For all other titles please Nuytsia 23, 589-606 \\KENS-file-001 contact [email protected] \Inmagic\FullTextFiles/080057-23.031.pdf Gosper CR, Prober SM, Yates CJ (2013). Multi- Margaret Byrne century changes in vegetation structure and fuel Director availability in fire-sensitive eucalypt woodlands. Science & Conservation Division Forest Ecology and Management 310, 102-109 \\KENS-file-001\Inmagic\StaffOnly/C19292.pdf Journal Publications Gosper C, Prober S, Yates C (2013). The role of Barrett RL, Lowrie A (2013). Typification and fire in plant conservation in wheatbelt remnants. application of names in Drosera section Australasian Plant Conservation 22, 21-22 Arachnopus (Droseraceae). Nuytsia 23, 527-541 \\KENS-file-001\Inmagic\StaffOnly/C19281.pdf \\KENS-file-001\Inmagic\FullTextFiles/080057- 23.029.pdf Gosper CR, Yates CJ, Prober SM (2013). Floristic diversity in fire-sensitive eucalypt woodlands Biggs LJ, Parker CL (2013). Updates to Western show a U-shaped relationship with time since Australia's vascular plant census for 2012. -

Rangelands, Western Australia
Biodiversity Summary for NRM Regions Species List What is the summary for and where does it come from? This list has been produced by the Department of Sustainability, Environment, Water, Population and Communities (SEWPC) for the Natural Resource Management Spatial Information System. The list was produced using the AustralianAustralian Natural Natural Heritage Heritage Assessment Assessment Tool Tool (ANHAT), which analyses data from a range of plant and animal surveys and collections from across Australia to automatically generate a report for each NRM region. Data sources (Appendix 2) include national and state herbaria, museums, state governments, CSIRO, Birds Australia and a range of surveys conducted by or for DEWHA. For each family of plant and animal covered by ANHAT (Appendix 1), this document gives the number of species in the country and how many of them are found in the region. It also identifies species listed as Vulnerable, Critically Endangered, Endangered or Conservation Dependent under the EPBC Act. A biodiversity summary for this region is also available. For more information please see: www.environment.gov.au/heritage/anhat/index.html Limitations • ANHAT currently contains information on the distribution of over 30,000 Australian taxa. This includes all mammals, birds, reptiles, frogs and fish, 137 families of vascular plants (over 15,000 species) and a range of invertebrate groups. Groups notnot yet yet covered covered in inANHAT ANHAT are notnot included included in in the the list. list. • The data used come from authoritative sources, but they are not perfect. All species names have been confirmed as valid species names, but it is not possible to confirm all species locations. -

Biodiversity Summary: Rangelands, Western Australia
Biodiversity Summary for NRM Regions Guide to Users Background What is the summary for and where does it come from? This summary has been produced by the Department of Sustainability, Environment, Water, Population and Communities (SEWPC) for the Natural Resource Management Spatial Information System. It highlights important elements of the biodiversity of the region in two ways: • Listing species which may be significant for management because they are found only in the region, mainly in the region, or they have a conservation status such as endangered or vulnerable. • Comparing the region to other parts of Australia in terms of the composition and distribution of its species, to suggest components of its biodiversity which may be nationally significant. The summary was produced using the Australian Natural Natural Heritage Heritage Assessment Assessment Tool Tool (ANHAT), which analyses data from a range of plant and animal surveys and collections from across Australia to automatically generate a report for each NRM region. Data sources (Appendix 2) include national and state herbaria, museums, state governments, CSIRO, Birds Australia and a range of surveys conducted by or for DEWHA. Limitations • ANHAT currently contains information on the distribution of over 30,000 Australian taxa. This includes all mammals, birds, reptiles, frogs and fish, 137 families of vascular plants (over 15,000 species) and a range of invertebrate groups. The list of families covered in ANHAT is shown in Appendix 1. Groups notnot yet yet covered covered in inANHAT ANHAT are are not not included included in the in the summary. • The data used for this summary come from authoritative sources, but they are not perfect. -

Development and Validation of Acacia Koa and A. Koaia Nuclear Ssrs Using Illumina Sequencing
Lawson and Ebrahimi ∙ Silvae Genetica (2018) 67, 20-25 20 Development and validation of Acacia koa and A. koaia nuclear SSRs using Illumina sequencing Shaneka S. Lawson1*, Aziz Ebrahimi2 1 USDA Forest Service, Northern Research Station, Hardwood Tree Improvement and Regeneration Center (HTIRC), Purdue University, Department of Forestry and Natural Resources, 715 West State Street, West Lafayette, IN 47907 2 Purdue University, Forestry and Natural Resources, Pfendler Hall, 715 West State Street, West Lafayette, Indiana 47907 * Corresponding author: Shaneka S. Lawson, Email: [email protected] Abstract loci. Some of the earliest uses of these technologies in plants are Morgante and Olivieri (1993) and Young (1996). These Koa (Acacia koa) and sub-species koaia (A. koaia) are two of markers provide effective means for investigating genetic more than 1,200 species from the genus Acacia within the diversity and accelerating genome studies. Construction and Leguminosae (also designated Fabaceae) family. In the past, development of SSRs provide a valuable resource for research- koa and koaia forests were found throughout the Hawaiian ers and are essential for subsequent studies of breeding and archipelago but populations have dramatically decreased. genetic diversity. The majority of marker-based research Comprehensive analyses of simple sequence repeats (SSRs) employed microsatellites for analysis of genetic differentiation have not been published previously. Here we use genome within koa and koaia populations (Fredua-Agyeman et al. 2008) sequencing and bioinformatics tools to report development of and amplified fragment length polymorphisms (AFLPs) or 100,000+ nuclear SSR (nuSSR) markers for use in koa and koaia quantitative reverse-transcriptase PCR (qRT-PCR) for pathoge- genome studies with transcriptome SSR information was inclu- nicity studies (Shiraishi et al. -

Floristic Survey/Inventory of Cane River Conservation Park
Vascular Flora of Cane River Conservation Park, Western Australia A report to the Bush Blitz Program, Australian Biological Resources Study Adrienne Markey1 & Steven Dillon2 1: Western Australian Conservation Science Centre, Department of Environment and Conservation, Kensington, Perth 2: Western Australian Herbarium, Department of Environment and Conservation, Kensington, Perth January 2012 © Crown copyright reserved Bush Blitz – Cane River Conservation Park, June 20 – 30th 2011 1 Nomenclature and taxonomy used in this report (as applicable) is consistent with that from: Florabase – The Western Australian Plant Census (WACensus) http://florabase.dec.wa.gov.au/ The Australian Plant Name Index (APNI) http://www.anbg.gov.au/databases/apni-about/index.html The Australian Plant Census (APC) http://www.anbg.gov.au/chah/apc/about-APC.html Family taxonomy follows the APG III system (Angiosperm Phylogeny Group 2009). Cover photo. View from a ridge of banded iron formation of surrounding Acacia bivenosa and Triodia hummock grasslands on stony plains, near Cane River homestead. Bush Blitz – Cane River Conservation Park, June 20 – 30th 2011 2 Contents Contents .....................................................................................................................................2 List of contributors.....................................................................................................................3 Abstract......................................................................................................................................4 -

Pasture Condition Guides for the Pilbara
Research Library Miscellaneous Publications Research Publications 10-2002 Pasture condition guides for the Pilbara A L. Payne A A. Mitchell Follow this and additional works at: https://researchlibrary.agric.wa.gov.au/misc_pbns Part of the Agricultural Education Commons, Biodiversity Commons, Environmental Education Commons, Environmental Indicators and Impact Assessment Commons, Environmental Monitoring Commons, Natural Resources and Conservation Commons, Natural Resources Management and Policy Commons, Soil Science Commons, and the Sustainability Commons Recommended Citation Payne, A L, and Mitchell, A A. (2002), Pasture condition guides for the Pilbara. Department of Primary Industries and Regional Development, Western Australia, Perth. Report 19/2002. This report is brought to you for free and open access by the Research Publications at Research Library. It has been accepted for inclusion in Miscellaneous Publications by an authorized administrator of Research Library. For more information, please contact [email protected]. Miscellaneous Publication 19/2002 ISSN 1326-4168 PASTURE CONDITION GUIDES FOR THE PILBARA Cover picture: Pump well pool, Mount Florance Station, looking south to the Hamersley Ranges Miscellaneous Publication 19/2002 Agdex 320/11 ISSN 1326-4168 PASTURE CONDITION GUIDES FOR THE PILBARA A.L. Payne Senior Adviser, Department of Agriculture, Western Australia and A.A. M itchell Former Adviser, Department of Agriculture, Western Australia (now c/- Department of Business, Industry and Resource Development, Darwin, Northern Territory) October 2002 © Director General, Department of Agriculture, Western Australia Pilbara Pasture Condition Guides The Director General of the Department of Agriculture and the State of Western Australia accept no liability whatsoever by reason of negligence or otherwise arising from the use or release of this information or any part of it. -

2014–153.52 Mb
DIRECTOR'S MESSAGE The past year has presented many challenges for the Division as we sought to meet the expectations of government at a time of fiscal constraint. These challenges presented the opportunity to review our activities to focus on the core functions we are required to deliver to support government priorities. Our Strategic Plan identifies how our science is integrated into delivery of the Department’s strategic goals of wildlife management, parks management, forest management, and managed use of natural assets. In reviewing our business we articulated our core functions as conserving threatened plant, animals and communities, landscape scale conservation, wildlife assets and inventory, WA Herbarium, bushfire research, marine monitoring and research, forest monitoring and research, wetland monitoring, hydrological monitoring, and data management and distribution. We have aligned our resources with these functions and refocused our activities to ensure we are delivering best practice science driven by management priorities to inform effective wildlife conservation and parks management. Through all of this we have continued to deliver significant scientific information to support conservation of our diverse plants, animals and ecological communities. Highlights of activities this year include publication of a book documenting the outcomes of the Kimberley Islands Biological Survey; documenting the response of mammals to the establishment of a patchy early dry season burning regime in the Kimberley; development of genetic monitoring -

Botanical.Survey of Central Pilbara Uplands
BOTANICAL.SURVEY OF CENTRAL PILBARA UPLANDS Project (N709) Progress Report 1999 Prepared by: Stephen van Leeuwen Date: May 1999 PRECIS BOTANICAL SURVEY OF CENTRAL PILBARA UPLANDS Proiect N706 The botanical survey of Central Pi/bara upland sites commenced in 1995. During the past four years 80 upland sites have been visited across the Hamersley Range. These sites extend from Pannawonica in the west to Newman in the east, from Wittenoom in the north to Pararburdoo in the south. The upland sites range in elevation from 600 m to the height of Western Australian's highest mountain, Mt Meharry at 1 250 m. On each of the sampled upland sites one 2.5 ha permanent benchmark quadrat was established. Within this quadrat all plant species present were recorded and voucher specimens collected for unknown taxa. Systematic intensive opportunistic flora sampling was also undertaken outside the quadrat over the entire upland site. A total of 265 person hours was accumulated in sampling upland sites. A total of 260 identified species have been recorded from these upland sites. These species represent 124 genera in 53 families. The identification of many other taxa remains undetermined. A total of 1 018 specimens represented by over 2 500 vouchers have been collected under the auspices of this project. Many species of conservation, biological and taxonomic significance have been recorded during this project. The majority of the significant species are represented by.disjunctly distributed populations and typically represent new records for the Pilbara Biogeographical Region. Preliminary, exploratory analyses of the specimen database and GIS atlas developed for this project indicate that elevation, bum history and edaphic (geology) considerations may be influencing the biogeographical patterning of upland species and plant communities across the Hamersley Range. -
Native Seed Transfer Zones in Australia How Far Can Seed Go?
NATIVE SEED TRANSFER ZONES IN AUSTRALIA HOW FAR CAN SEED GO? JUNE 2021 First published 2021 Project Phoenix Greening Australia (National Office) Level 3, 349 Collins Street Melbourne VIC 3000 Tel: 1300 886 589 Email: [email protected] Website: www.greeningaustralia.org.au ISBN: xxx-x-xxxxxx-xx-x (Book) xxx-x-xxxxxx-xx-x (epub) Authors: Dr Nola Hancock, Department of Biological Sciences, Macquarie University Dr. Francisco Encinas-Viso, The Centre for Australian National Biodiversity Research and CSIRO Title: Native seed transfer zones in Australia — How far can seed go? Notes: Includes bibliographical references Copyright © Project Phoenix 2021 Cover by Kerry O’Flaherty, Design Consultant Internal design by Puddingburn Publishing Services Proofread by Puddingburn Publishing Services This report is copyright. Except for private study, research, criticism or reviews, as permitted under the Copyright Act 1968 (Cth), no part of this report may be reproduced, stored in a retrieval system or transmitted in any form or by any means without prior written permission. Enquiries should be made to [email protected]. Project Phoenix is supported by the Australian Government’s Wildlife and Habitat Bushfire Recovery program and co-ordinated by Greening Australia. Across all of our Project Phoenix activities and actions we pay respect to the Traditional Owners and Custodians of the lands and waters on which we work. We honour the resilience and continuing connection to country, culture and community of all Aboriginal and Torres Strait Islander people across Australia. We recognise the decisions we make today will impact the lives of generations to come. ACKNOWLEDGEMENTS The authors gratefully thank and acknowledge the assistance all interviewees gave to this project.